A method for rapid identification of chromosomal ploidy in Avena plants and its application
A chromosome ploidy, plant technology, applied in the field of plant biology, can solve the problems of high cytology operation technology, difficult large-scale quantification, time-consuming and labor-intensive
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0032] The extraction of embodiment 1 Avena genome DNA
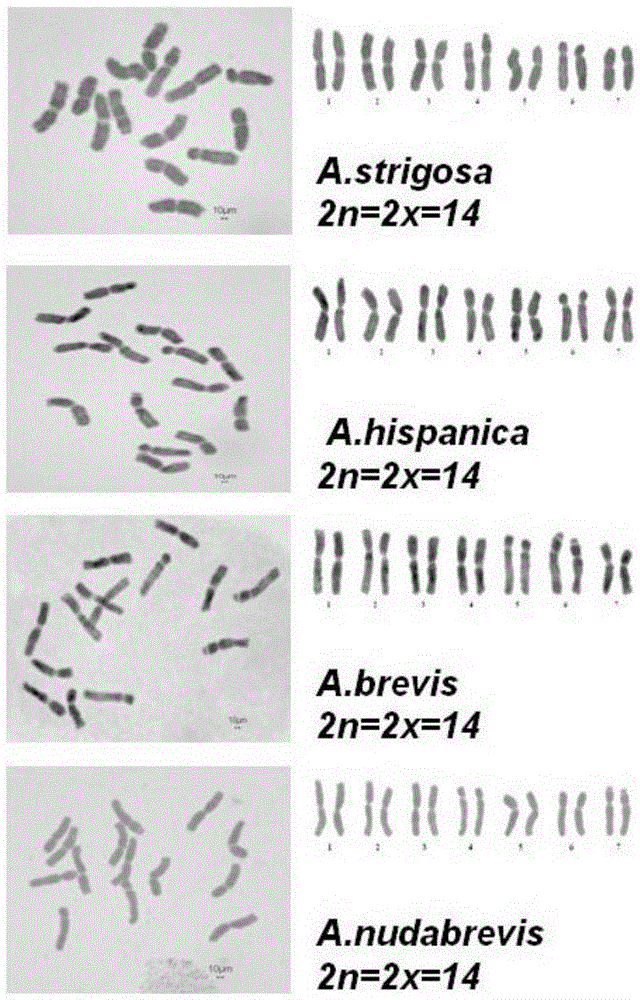
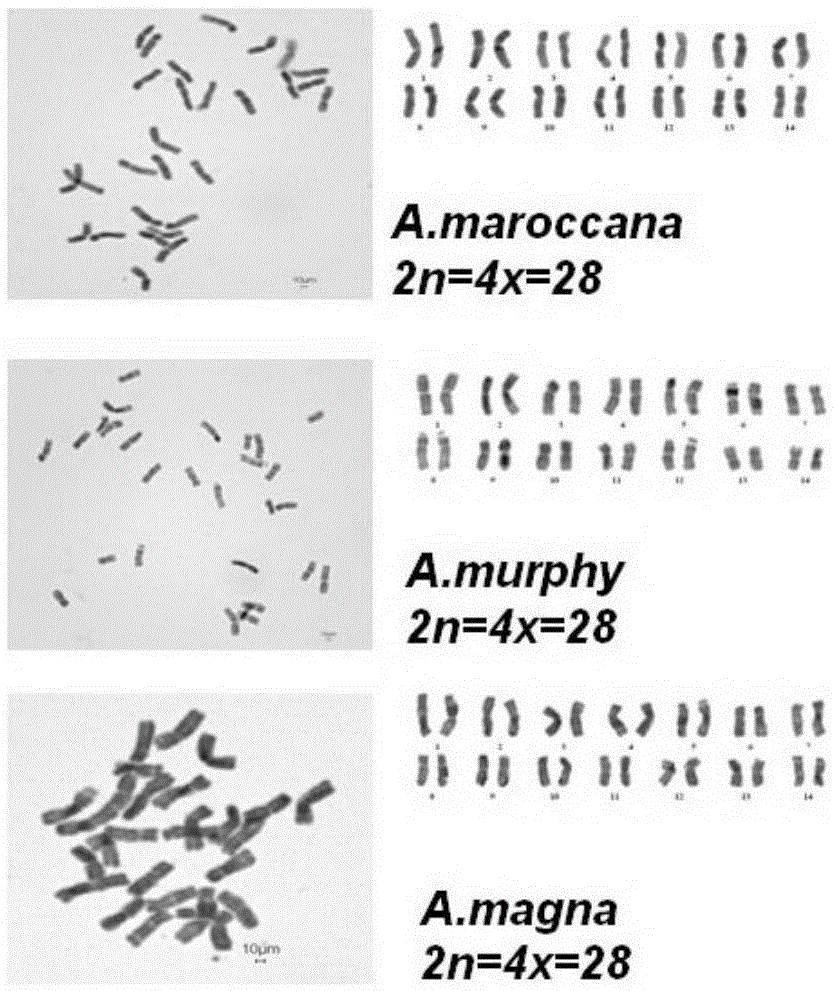
[0033] In this embodiment, A.strigosa (sand oat), A.hispanica (Spanish oat), A.brevis (short oat), A.nudabrevis (small grain naked oat), A.maroccana (Maloca oat), A.murphy (Murphy oats), A.magna (large oats), A.sativa (common cultivated oats), A.fatua (common wild oats), A.sterilis (wild red oats) and A.chinensis (naked oats ) as the experimental object.
[0034] Using the young leaves of the above-mentioned plants of the genus Avena as materials, oat DNA was extracted by using the CTAB method. The specific method is as follows: take 0.1 g of the material, add liquid nitrogen and grind it quickly. Transfer the ground material into a 1.5ml centrifuge tube, immediately add 600μl of preheated 2×CTAB buffer solution, mix by inverting, keep warm at 65°C for 10-20min, and shake it from time to time. Add an equal volume of chloroform / isoamyl alcohol (volume ratio 1 / 1, the same below), mix gently by inversion, and centrifuge a...
Embodiment 2
[0035] Embodiment 2 to the PCR amplification of Avena plant genomic DNA
[0036] Various oat genome DNAs extracted in embodiment 1 are carried out PCR amplification, and method is as follows:
[0037] Take a clean and sterile 0.2ml centrifuge tube, add the following reagents to it: ddH 2 O, 36 μl; 10×PCRBuffer, 5 μl; dNTP (10 mMeach), 1 μl; upstream primer (SP1) / downstream primer (SP2) of a specific primer pair, 2 μl; oat genomic DNA, 0.1-1 μg; Taq enzyme, 1 μl.
[0038] The nucleotide sequence of the specific primer pair is:
[0039] Upstream primer (SP1): 5'-AGCGTCTGCTTCAAAATCTGTT-3' (as shown in SEQ ID NO.1)
[0040] Downstream primer (SP2): 5'-TTTCTTCCTGCCGCGTTAAGTT-3' (shown in SEQ ID NO.2).
[0041] Amplification was carried out according to the following reaction conditions: 94°C for 5 min; 94°C for 20 sec, 55°C for 30 sec, 72°C for 1 min, 35 cycles; 72°C for 7 min.
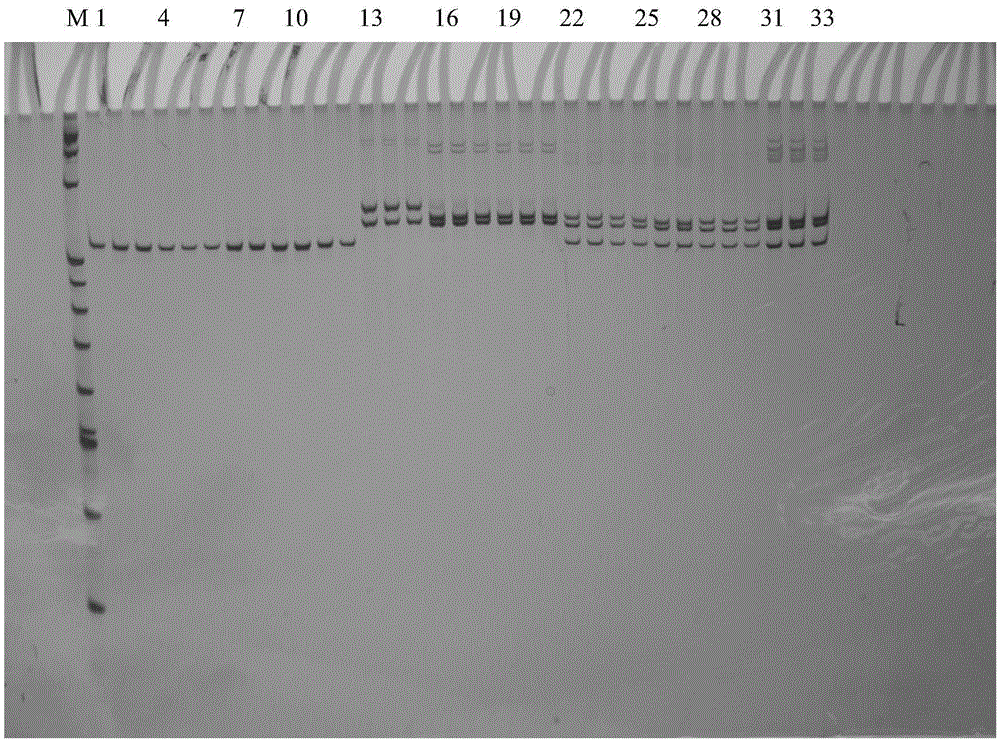
[0042] The polyacrylamide gel electrophoresis detection of embodiment 3 amplification result
[00...
Embodiment 4
[0049] Example 4 Observation and Identification of Chromosomal Ploidy in Avena Plants
[0050] With reference to methods such as Liu Wei [Liu Wei, Zhang Zongwen, Wu Bin. Karyotype identification of the diploid oat germplasm introduced in Canada. Journal of Plant Genetic Resources. 2013,14(1):141-145], for example 1- 3 The chromosome ploidy of the Avena genus involved is observed and identified, and the specific methods are as follows:
[0051] Take oat seeds with full grains and put them in a clean petri dish, add water to submerge the seeds, and place them in a constant temperature incubator at 23°C to cultivate until they are white, then put the white seeds in a clean petri dish lined with sterilized water-soaked filter paper, and place Put it in the refrigerator at 4°C for 48 hours, and then put it in a constant temperature incubator at 23°C to grow roots. When the roots grow to 1.0-2.0cm, cut the new and strong root tips and pretreat them in ice water for 48 hours, and the...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com