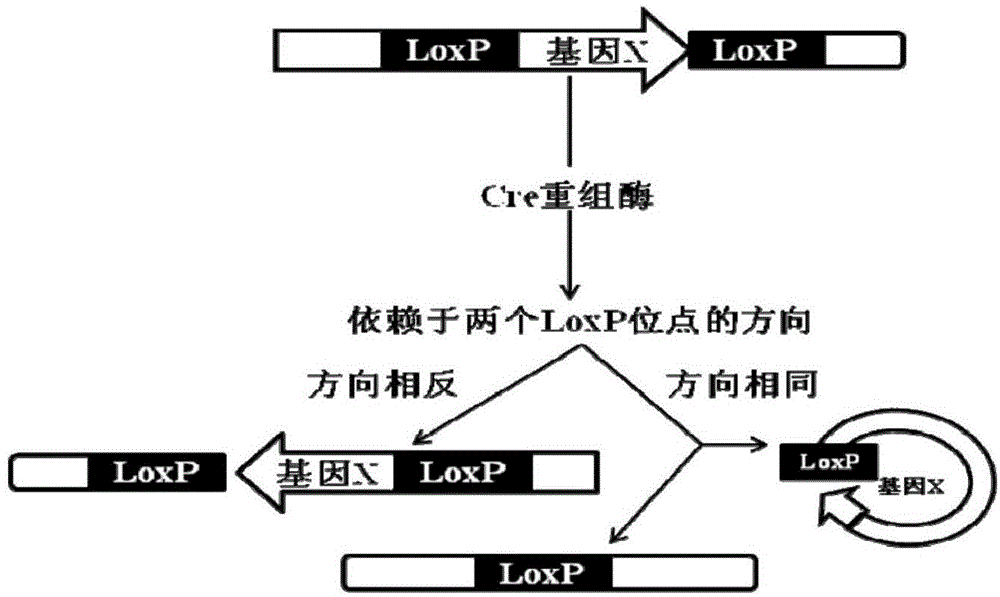
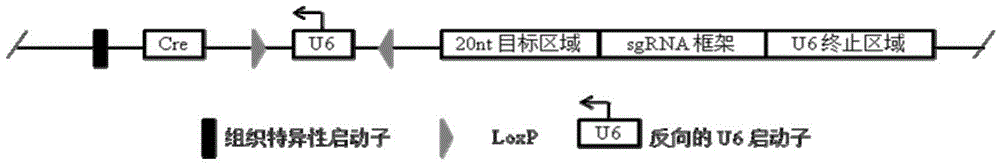
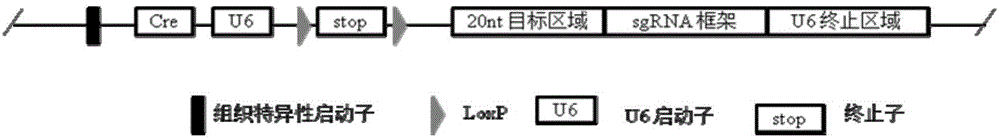
Conditional gene knockout method based on CRISPR/Cas9 technology
A gene knockout and positive technology, applied in the field of gene modification, can solve the problems that tissue, cell, and space-time specific knockout cannot be achieved, it is difficult to obtain double transgenic offspring mice, and the identification and screening process is complicated, reaching the test cycle Short, reduced cytotoxicity, short cycle effect
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0074] 1. Experimental materials
[0075] ① HD Clinging Kit (639648): purchased from Clontech;
[0076] ②Premix Taq (D331A), Primerstar (DRO44A), dNTP (4030Q): purchased from Yubao Bioengineering (Dalian) Co., Ltd. (Takara);
[0077] ③ Restriction endonucleases BbsI (R0539L), AsisI (R0630L), HpaI (R0105S): purchased from Beijing Bomeisi Biotechnology Co., Ltd. (NEB); Church Cloning Vector is a commercial plasmid, purchased from Church through Addgene laboratory; pCR-Blunt II-TOPO is also a commercial plasmid, purchased from Invitrogen through Addgene; plasmid with Cre recombinase: purchased from Albee Messing Laboratories through Addgene; source of Rosa26 expression vector, purchased from Addgene at Liqun Luo's lab.
[0078] 2. Gene structure and sequence analysis
[0079] Target gene: mouse eukaryotic translation initiation factor 3 (Eif3h gene);
[0080] Ensembl gene code number: ENSMUSG00000022312;
[0081] Eif3h gene structure: Eif3h gene contains 8 exons, including...
Embodiment 2
[0113] Example 2 Construction of Cas9 expression vector and production of Cas9 tool mouse
[0114] (1) Amplification of Cas9 protein
[0115] ① Design primers for amplifying Cas9 protein, as follows:
[0116] Cas9-F: CGCGGTCTTTCCAGTGATCGATTAGTTATTAATAGTAATCAA
[0117] Cas9-R: CTCTAGTCCGCGGGTGCGATAGCTCACACCTTCCTCTTCTTTCTTG
[0118] ②PCR amplified the full sequence of Cas9 protein, the system is as follows:
[0119]
[0120] The PCR program is as follows:
[0121]
[0122]
[0123] ③ PCR products were subjected to agarose gel electrophoresis, such as Figure 9 shown.
[0124] (2) Cas9 protein connected to Rosa26 carrier
[0125] ① Use Clontech's The HD Clinging Kit will detect the correct Cas9 sequence and connect it to the vector linearized by AsisI and HpaI enzymes. The vector is transformed by the company, and it has a broad-spectrum expression promoter CMV, the 5'arm of Rosa26, and the Rosa26 vector backbone of the 3'arm. The map is as follows Figure 10 s...
Embodiment 3
[0137] Example 3 Obtaining conditional knockout mice
[0138] The Cas9 tool mouse was crossed with the sgRNA mouse, and finally 15 mice were obtained. Different tissues of the mice were taken, and the genome was extracted. After PCR amplification with Eif3h-F / Eif3h-R primers, they were sent to Suzhou Jinweizhi Co., Ltd. Sequencing results showed that the gene knockout rate in the liver tissue reached over 80%, while the genes in other tissues remained intact.
[0139] The above primer pair: Eif3h-F:ATCATATATTTAATTTTCAACAAGT
[0140] Eif3h-R: CTTTCCTACAGAGCTTCACCT
[0141] Analysis of gene knockout results:
[0142] liver tissue:
[0143]
[0144] Wild type refers to the liver tissue of mice without any modification;
[0145] Samples 1 to 15 refer to experiments on different tissues of 15 mice. The genomes of different tissues were extracted, amplified by PCR, and then sent for sequencing. The results showed that only the Eif3h gene of the liver tissue was modified while...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com