LRR-RLK (leucine-rich repeat receptor-like kinase) gene in arachis hypogaea.L and application thereof to bacterial wilt resistance of tobaccos
A peanut and genetic technology, applied in the fields of application, genetic engineering, plant genetic improvement, etc., can solve the problems of peanut bacterial wilt, threat to peanut yield and quality, negative correlation between resistance and high yield and high quality, and achieve important application value, Effect of Improving Resistance to Bacteria Wilt
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
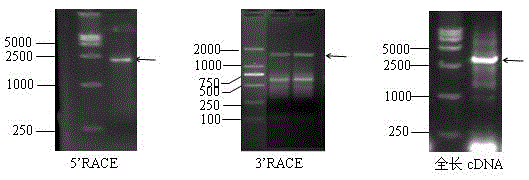
[0017] [Example 1] RACE acquisition AhRLK6 Gene 3' Unknown Sequence and 5' Unknown Sequence
[0018]Based on peanut abiotic and biotic stress 454 sequenced transcripts, the peanut expression profile gene chip (synthesized by Roche Company) was combined, and the candidate gene fragments were obtained by hybridization of the chip hybridization before and after inoculation of R. solanacearum strains before and after induction of R. solanacearum. Design a pair of gene primers PRRS_6_F (5'-ACGAGATTCGCTTCATGACGAGCCTC-3') and PRRS_6-R (5'-TCTAAGCATTCCTACCCACCTCCTCTG-3'); then add the linker sequence RACE-F (AAGCAGTGGTATCAACGCAGAGTGGCCAT) and RACE-R (ATTCTAGAGGCCGAGGCGGCCGACATGd(T)30N-1N-3'), using PRRS_6-R primers and RACE-F primers and PRRS_6_F primers and RACE-R primers for 5' and 3'- RACE reactions, respectively, 5'- RACE reaction conditions are 94°C 5min→(94°C 30s→57°C 30s→72°C 2min) 30 cycles→72°C 10min; 3′-RACE reaction conditions are 94°C 5min→(94°C 30s→72°C 2min) 5cycles→ (...
Embodiment 2
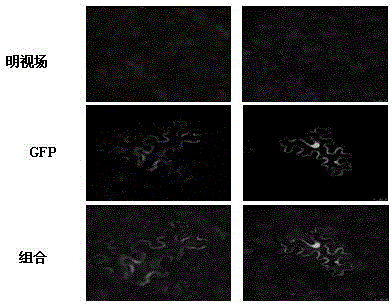
[0019] [Example 2] AhRLK6 Subcellular localization of gene expression products
[0020] For the subcellular localization vector, amplify from the plasmid with complete reading frame by AhRLK6-SL-F (5'-ATTAGGATCCACCATGAGAACCTCATTGTGTTGTAG-3') and AhRLK6-SL-R (5'-ATTAGGCGCGCCAGAGATTAATTAGGTTATGATGGGT-3') primers get excluding the kill password AhRLK6 Gene cDNA, with BamH1 and Asc1 restriction sites at the 5' end and 3' end respectively, for the pBI-GFP constructed in our laboratory and the amplified AhRLK6 The gene was double digested with BamH1 (purchased from NEB Company) and Asc1 (purchased from NEB Company) at the same time, and p35S was constructed after ligation and transformation:: AhRLK6 ::GFP vector. Agrobacterium GV3101 was transformed by liquid nitrogen freeze-thaw method, and p35S::GFP was used as a control, and Agrobacterium infiltration method was used to transform Nicotiana benthamiana. After culturing at 25°C for 48 hours, the green fluorescence was observed w...
Embodiment 3
[0021] [Example 3] AhRLK6 Analysis of expression patterns after treatment with R. solanacearum resistant and susceptible to R. solanacearum
[0022] In order to analyze the difference in the expression of AhRLK6 gene in bacterial wilt resistant and susceptible peanut varieties inoculated with R. XH) After inoculation of R. solanacearum by leaf cutting method, the expression of the gene was analyzed at different time points. The total RNA of peanut and tobacco leaves was extracted by the CTAB method, and single-stranded cDNA was synthesized by reverse transcription according to the instructions of PrimeScript Reverse Transcriptase reverse transcriptase (purchased from TAKARA Company). The single-stranded cDNA was diluted 10 times, 2 μL was used as a template, and Ahactin, Ntactin As an internal reference gene, Eppendorf Mastercycler ep realplex real-time fluorescent quantitative PCR instrument was used for quantitative PCR reaction. According to the reaction system of TAKARA S...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com