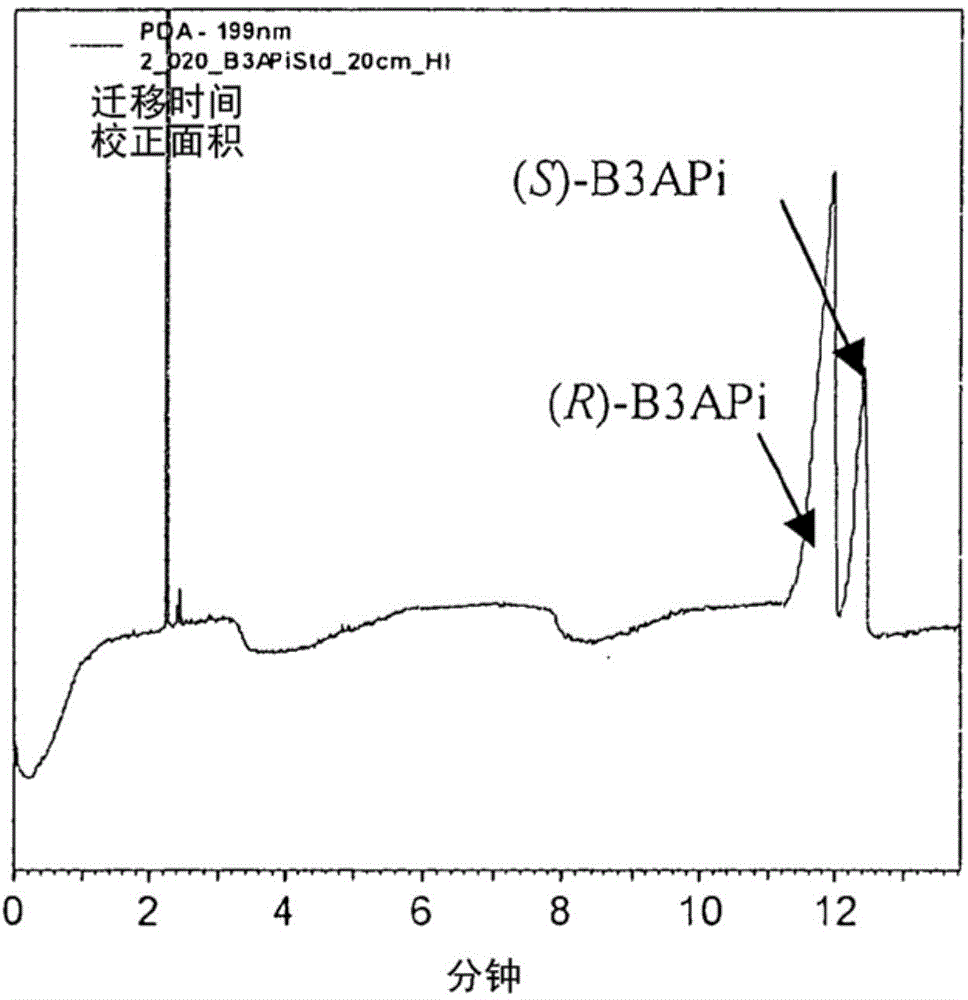
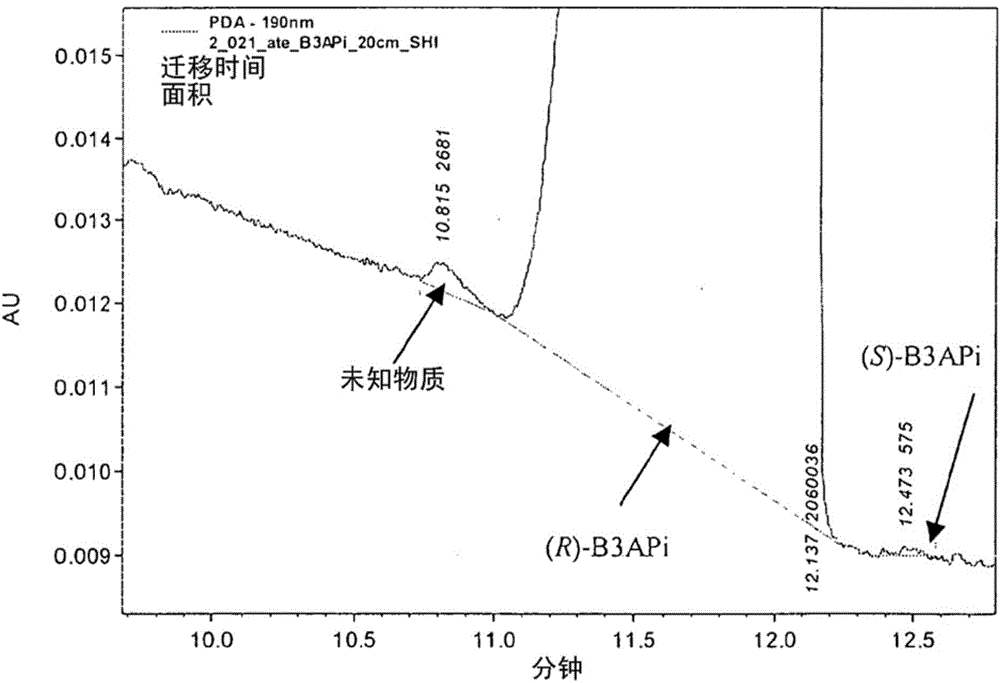
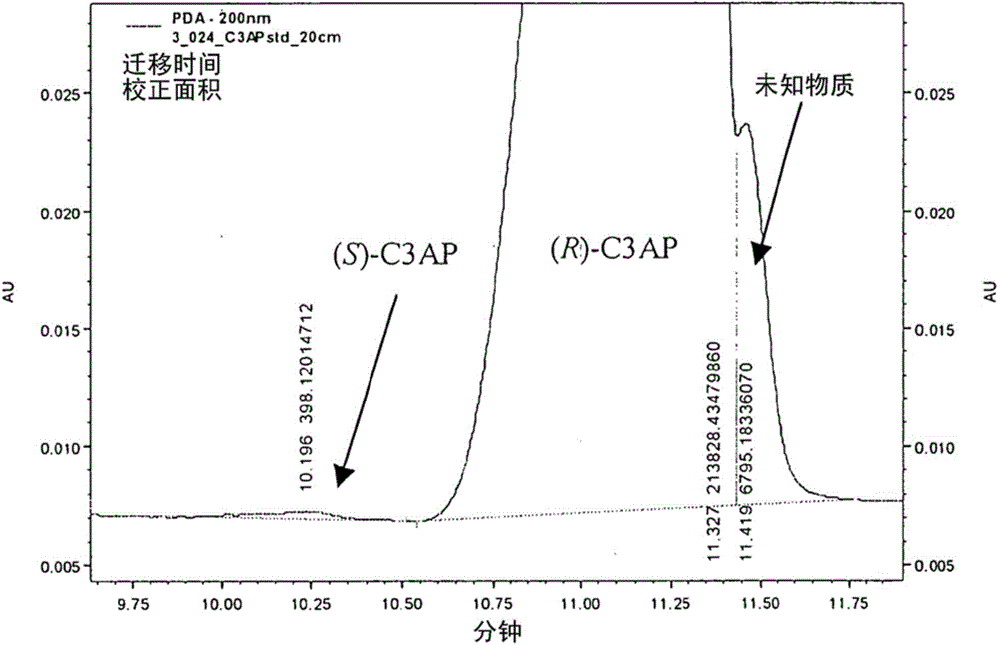
A process for the identification and preparation of a (r)-specific omega-transaminase
A technology for transaminase and amino, which is applied in the field of identification and preparation of (R)-specific ω-transaminase, which can solve the problems of unobtainable and economically feasible methods
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0154] Embodiment 1-identification of (R)-selective ω-transaminase
[0155] The amino acid sequence (SEQ ID no.5) of the (R)-selective ω-TA of Mycobacterium aureus given in EP 1038953A1 and the (R)-selective ω-TA of Arthrobacter given in EP 0987332A1 The amino acid sequence (SEQ ID no. 7) was used as the query biomolecule sequence. The biomolecule library used in this embodiment is the pubmed protein database of NCBI ( http: / / www.ncbi.nlm.nih.gov / pubmed (July 13, 2009).
[0156] The amino acid sequence (SEQ ID no.5 or 7) from Mycobacterium aureus or Arthrobacter ω-TA has been used as query sequence, using standard parameters (BLOSUM62 scoring matrix, word length: 3, gap penalty: presence-11, Extension-1) A BLAST search is performed to identify a first set of 100 different amino acid sequences from different organisms, all of which have a minimum of 30% sequence identity to the query sequence.
[0157] BLAST was performed on July 13, 2009 using "non-redundant protein sequen...
Embodiment 2-
[0162] Example 2- Preparation and analysis of transaminases from Aspergillus terreus, Mycobacterium vanbaalenii and Rhizobium japonicus
[0163] 2.1 ω-TA from Mycobacterium vanbaalenii (entry 9 in Table 1, SEQ ID no.49 and 50), ω-TA from Aspergillus terreus (entry 1 in Table 1, SEQ ID no.3 and 4) have been obtained , omega-TA from Rhizobium japonicum (entry 11 in Table 1, SEQ ID nos.1 and 2), and used (for Escherichia coli) in the form of codon usage (codon usage) suitable for use in Expression of suoshu enzyme in Escherichia coli. Hereinafter the Mycobacterium va nbaalenii transaminase is referred to as Mva-TA, the Aspergillus terreus transaminase as Ate-TA and the Rhizobium japonicum transaminase as Mlo-TA.
[0164] 2.2 Acetophenone test
[0165] Mva-TA and Ate-TA convert (R)-α-MBA faster than the (S)-enantiomer in the acetophenone assay using 2.5 mM amine and 2.5 mM pyruvate at pH 7.5 and 30°C At least 100 times.
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com