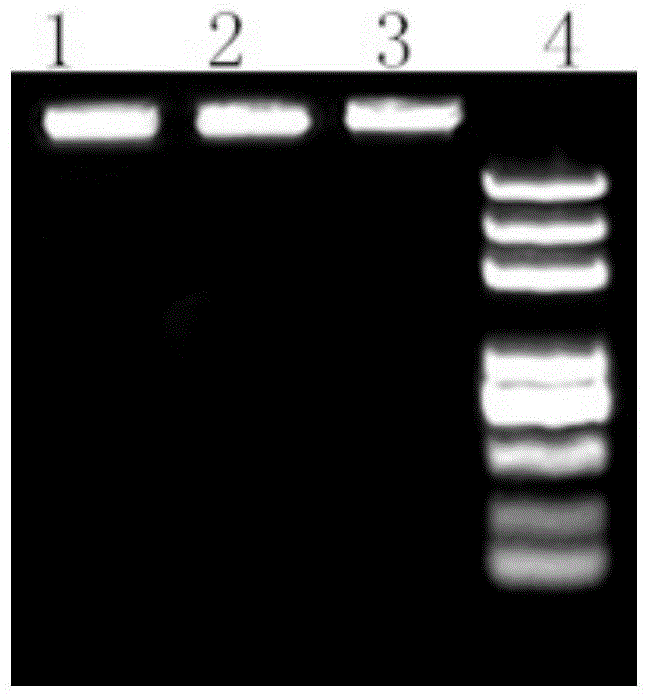
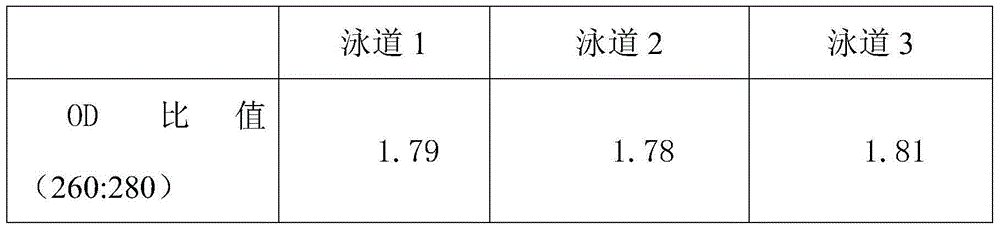
Method for rapidly extracting DNA of gram-positive bacterium genome based on paramagnetic particle method
A gram-positive bacteria and genome technology, applied in the biological field, can solve the problem of long extraction time, achieve the effects of economical extraction, simplify experimental steps and save extraction time
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0043] A method for rapidly extracting the genomic DNA of gram-positive bacteria based on a magnetic bead method, specifically comprising the following steps:
[0044] 1) Breakage of Gram-positive bacterial cells: Take 1ml of Staphylococcus overnight culture solution into a 1.5ml centrifuge tube, centrifuge at 12000rpm / min for 1min, discard the supernatant, add 1ml Staphylococcus overnight culture solution to the centrifuge tube, 12000rpm / min After centrifuging for 1-5 min, discard the supernatant, add 1ml of Staphylococcus overnight culture solution to the centrifuge tube, centrifuge at 12000rpm / min for 1min, discard the supernatant, (i.e. take 3ml of Staphylococcus overnight culture solution in total, and add in 3 times) into a 1.5ml centrifuge tube, centrifuge at 12000rpm / min for 1min each time and discard the supernatant), then add 0.25ml of double-distilled water to the bacterial pellet to suspend the pellet, and add 75μl of working solution A, incubate at 40°C for 30min, ...
Embodiment 2
[0058] A method for rapidly extracting the genomic DNA of gram-positive bacteria based on a magnetic bead method, specifically comprising the following steps:
[0059] 1) Breakage of Gram-positive bacterial cells: Take 1ml of Streptococcus overnight culture solution into a 1.5ml centrifuge tube, centrifuge at 10000rpm / min for 5min, discard the supernatant, add 1ml of Streptococcus overnight culture solution to the centrifuge tube, 10000rpm / min After centrifuging for 5 min, discard the supernatant, add 1ml of streptococcus overnight culture solution to the centrifuge tube, centrifuge at 10000rpm / min for 5min, discard the supernatant, (i.e., take 3ml of streptococcus overnight culture solution, and add it in 3 times. into a 1.5ml centrifuge tube, centrifuge at 10000rpm / min for 5min each time and discard the supernatant), then add 0.2ml of double distilled water to the bacterial pellet to suspend the pellet, and add 50μl of working solution A, incubate at 40°C for 10min, then add ...
Embodiment 3
[0072] A method for rapidly extracting genomic DNA of gram-positive bacteria based on a magnetic bead method, specifically comprising the following steps:
[0073] 1) Breakage of Gram-positive bacterial cells: Take 1ml of actinomycetes overnight culture solution into a 1.5ml centrifuge tube, centrifuge at 13000rpm / min for 2min, then discard the supernatant, add 1ml of actinomycetes overnight culture solution to the centrifuge tube, After centrifuging at 13000rpm / min for 2min, discard the supernatant, add 1ml of actinomycetes overnight culture solution to the centrifuge tube, discard the supernatant after centrifuging at 13000rpm / min for 2min, (that is, take a total of 3ml of actinomycetes overnight culture solution, divide into 3 Add it to a 1.5ml centrifuge tube once, centrifuge at 13000rpm / min for 2min each time and discard the supernatant), then add 0.5ml double distilled water to the bacterial pellet to suspend the pellet, add 80μl working solution A, and keep warm at 40°C ...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com