Brown tide algae species detection method and kit
A technology for detection kits and detection methods, applied in the direction of microorganism-based methods, biochemical equipment and methods, measurement/inspection of microorganisms, etc., can solve problems such as difficulties in identifying brown tide algae species, and achieve short detection time and easy control , good specific effect
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0093] A. anophagefferens pure algal strain sample to the verification of the constructed quantitative PCR detection method:
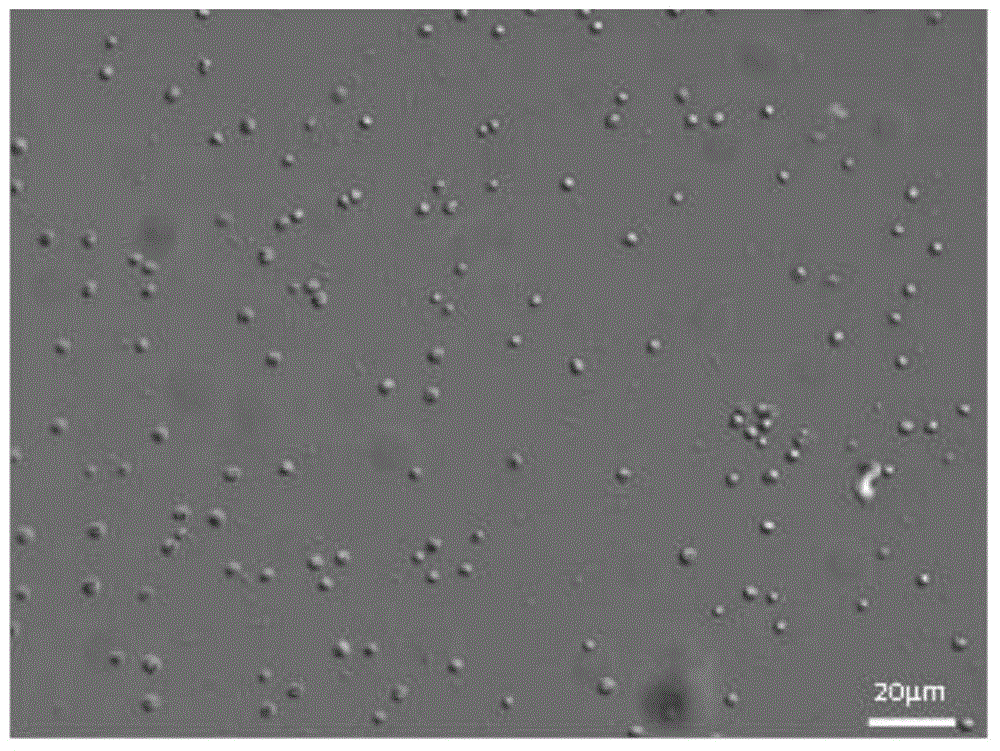
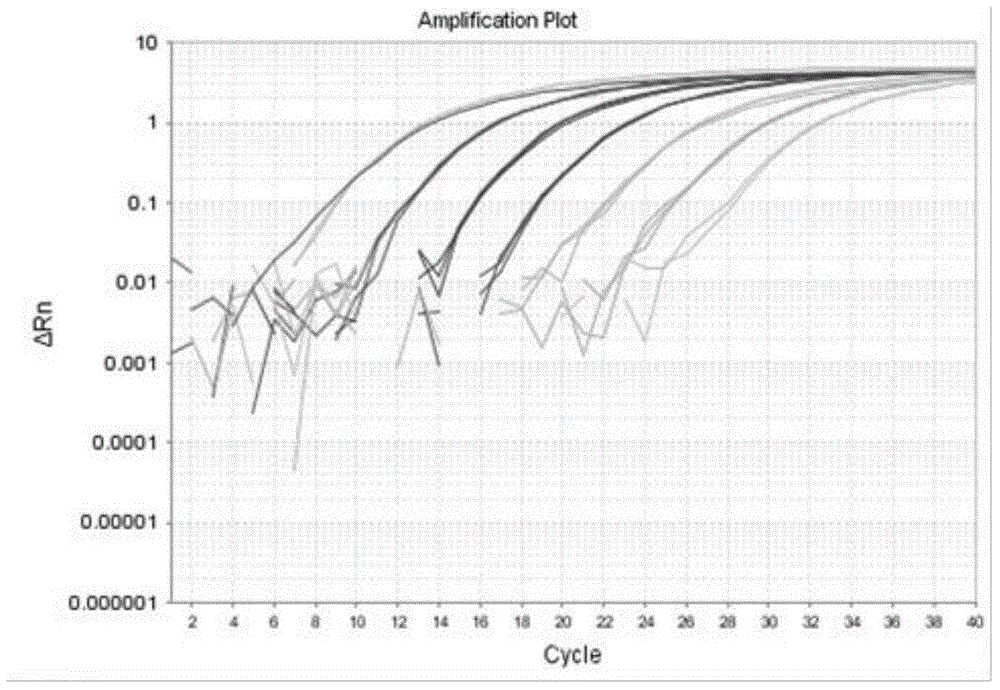
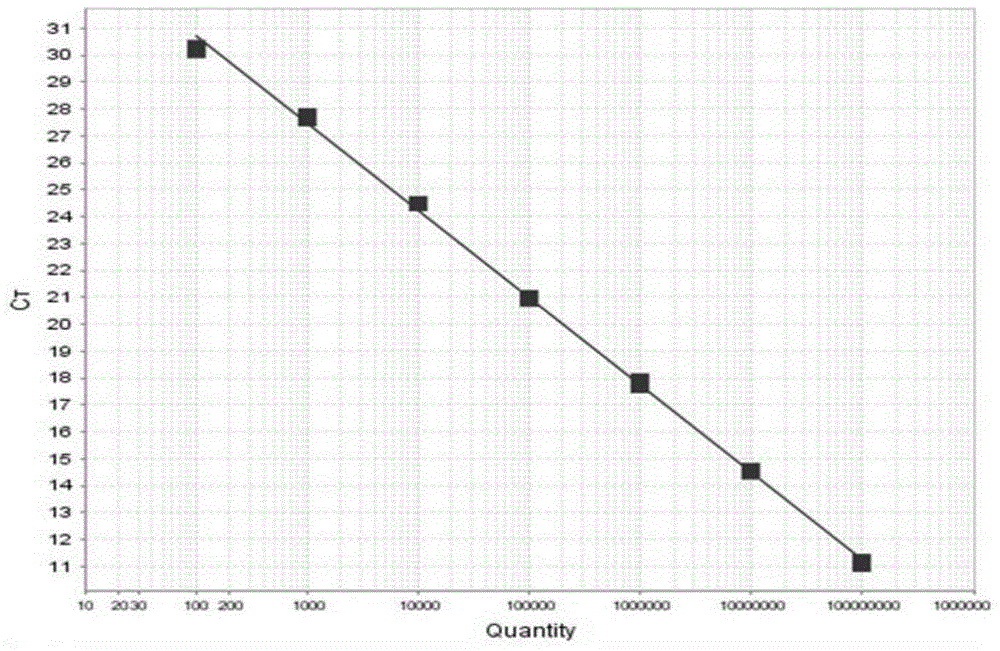
[0094] Take 3 sterilized 3000mL Erlenmeyer flasks, add 1200mL sterilized seawater, add (f / 2-Si) culture solution at a ratio of 1 / 1000, and take 300mL from the same culture flask of A.anophagefferens in the exponential growth phase Inoculate in these 3 Erlenmeyer flasks, after culturing for 9 days, count under the microscope, the counting method is the same as the step b) of the above-mentioned step 2), and the result is 3.2 × 10 6 cells / L. Filtered with a 0.45 μm filter membrane, the number of algae cells obtained was 3.2×10 6 cells, 3.2×10 3 cells and 3.2×10 2 Cells samples, and extract the DNA in these samples. After purification, use the purified DNA as a template to perform fluorescent quantitative PCR amplification with primers AanF, AanR and probe Aan probe. For the fluorescent quantitative PCR reaction system and reaction conditions, see The...
Embodiment 2
[0099] Embodiment 2 Utilizes the method of the present invention to detect environmental samples
[0100] First, sample collection: According to the main areas of brown tide outbreaks in Qinhuangdao's coastal waters from 2009 to 2012, the sampling station Q (119°'25.568''E, 39°'45.056'N) was set up, and the sampling time was from April 2013 to In September, the sampling frequency is once a month, collected at the beginning of the month. During the sampling period, the water temperature was 9.2-26.8°C and the salinity was 30-32psu.
[0101] Secondly, the extraction of total DNA from environmental samples: collect surface water samples (0-5cm) at station Q, take 500mL, filter it with a 0.45μm filter membrane immediately, and store the filter membrane (with algae cells) at -20°C for three times each time. in parallel; using the soil DNA extraction kit UltraClean TM SoilDNA Isolation Kit (Mo Bio Laboratories, Inc., Solana Beach, California, USA) extracted the total DNA of the a...
Embodiment 3
[0105] Example 3 Verification of the validity of the environmental sample test results:
[0106] In order to further verify the effectiveness of the A. anophagefferens algae cell fluorescent quantitative PCR detection technology described in the present invention in detecting environmental samples, high-throughput sequencing technology was used to simultaneously detect these environmental samples.
[0107] High-throughput sequencing technology (pyrosequencing), called next-generation sequencing technology, can not only detect various known micro-organisms, but also discover new species, so as to understand the whole picture of all micro-organisms in an environment, even those with a small amount. species. At present, this technology has been widely used to detect the composition of microbiological communities in various environmental samples, such as seawater, soil, and wastewater treatment systems.
[0108] The Qinhuangdao Q site obtained in Example 2 was used as a template ...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com