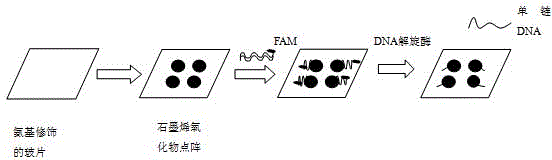
DNA helicase detection method based on graphene oxide chip
A DNA helicase, graphene oxide technology, applied in the field of biomedicine, can solve the problems of high cost and low detection throughput
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment Construction
[0019] The present invention will be further described below in conjunction with the accompanying drawings and specific embodiments, but the protection scope of the present invention is not limited thereto.
[0020] (1) Preparation of amino-modified glass slides: Add 5 ml of hydrogen peroxide (H 2 o 2 ), then
[0021] Then add 15 ml of concentrated sulfuric acid (H 2 SO 4 ), shaking slowly on a shaker or standing still for 30 min to make it
[0022] Fully react, this reaction can make the surface hydroxylated, pour off the liquid of the previous step reaction, wash 3 times with deionized water. First wash with absolute ethanol twice, then pour 20 ml of absolute ethanol, the obtained hydroxylated
[0023] The silicon wafer was transferred to a beaker and washed 3 times with absolute ethanol. Pour off the ethanol after cleaning, and quickly add a mixture of 3-aminopropyltriethoxysilane (APTES) and absolute ethanol (volume ratio 1:15), or add 15ml of absolute ethanol first,...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com