Kit for quantitatively detecting Ustilaginoidea virens
A technology for the quantitative detection of rice false smut bacteria, applied in the biological field, can solve the problems of no specific morphological characteristics, small rice smut spores, slow growth rate, etc.
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
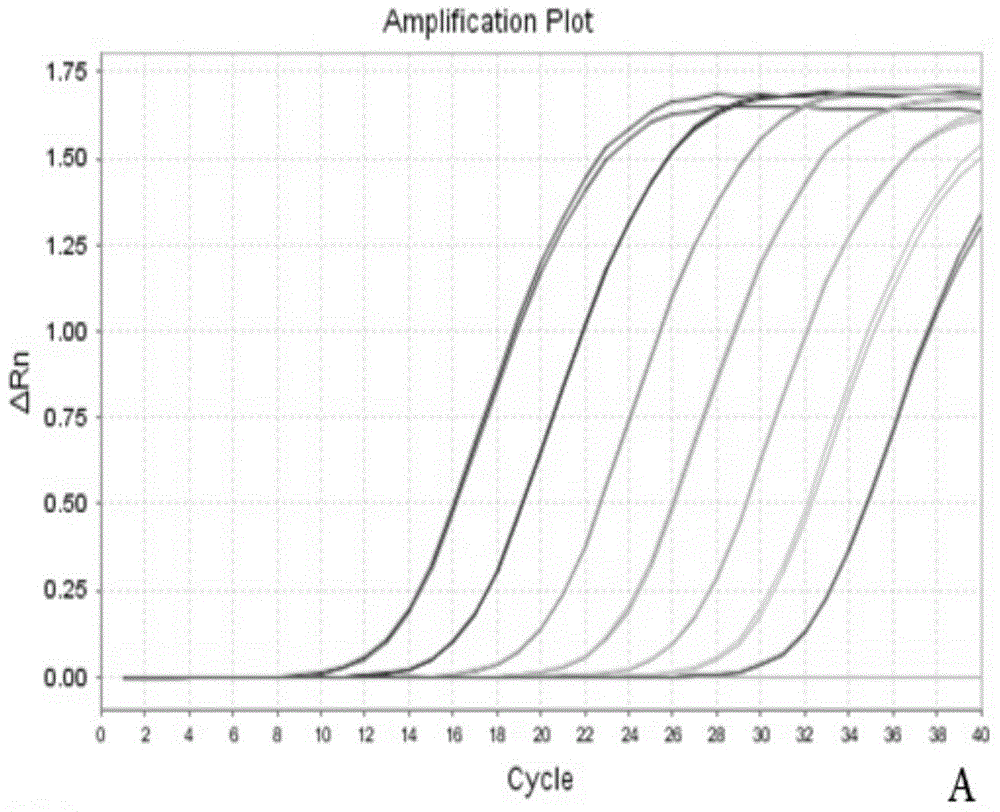
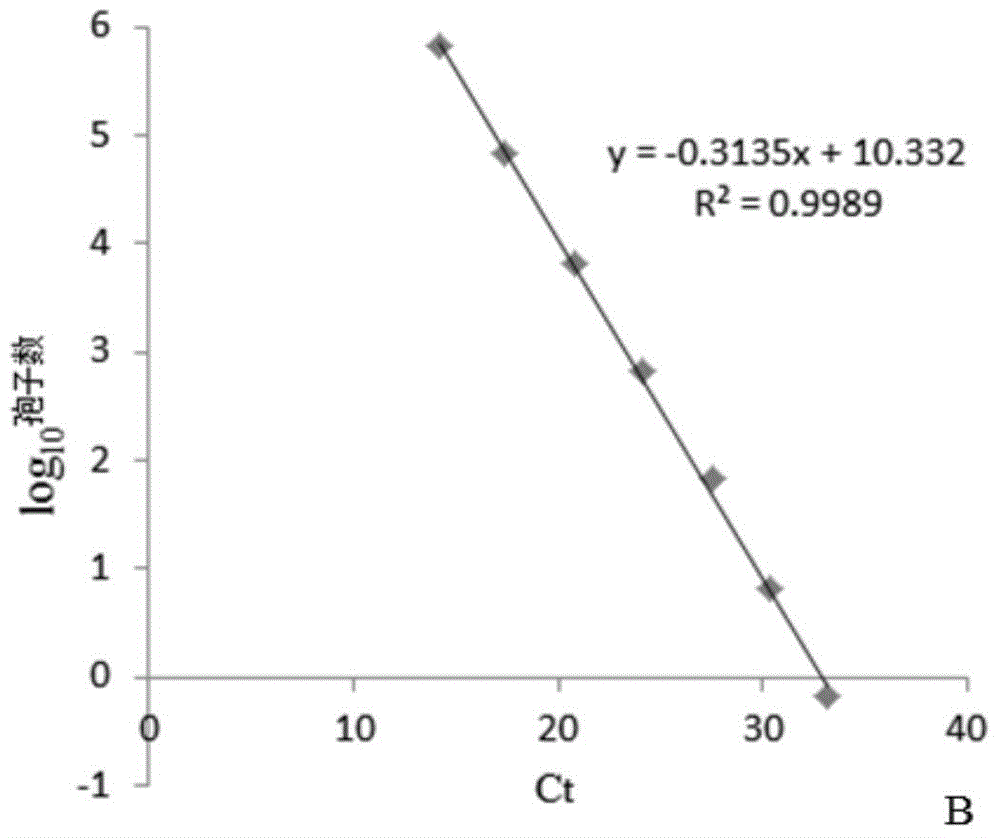
[0016] Embodiment 1: the spore quantitative detection of rice false smut
[0017] Genome extraction of spore suspension: Dilute the conidia of rice mistletoe with TE solution to obtain 4.2×10 7 , 4.2×10 6 , 4.2×10 5 , 4.2×10 4 , 4.2×10 3 , 4.2×10 2 , 4.2×10 / mL spore solution. Genomic DNA was extracted by taking 400 μL of the spore suspension, and finally obtaining 50 μL of each DNA sample solution. Take 2 μL respectively as a template for real-time PCR quantitative detection.
[0018] Oryza spp. spore DNA extraction: Add 0.4 g of pickled beads to each sample, put it in a Fast-prep instrument, process the sample for 40 s at a speed of 6 m / s, and repeat three times. Add 400 μL of DNA extraction buffer (10mM Tris HCL, pH7.5; 0.1mM EDTA, 6% SDS, 2% β-mercaptoethanol), mix well, and place in a 65°C water bath for 1 hour. Add 800μL of chloroformphenol (1:1), mix well, centrifuge at 12000g for 10min, and transfer the supernatant to a new centrifuge tube. Add 1 μL of glycogen...
Embodiment 2
[0021] Example 2: Quantitative detection of samples captured by airborne spores
[0022] Quantitative detection of spores of rice false smut in the spore capture samples from August 21 to September 10, 2012 in the rice-growing area of Ninghai County, Zhejiang Province. The 7-day spore capture instrument of Burkard Company was used for spore capture, and the polyester film was divided into small pieces according to the day, and loaded into a centrifuge tube with 400 μL of TE.
[0023] DNA extraction: Add 0.4 g of acid-washed beads to each sample, place in the Fast-prep instrument, process the sample for 40 s at a speed of 6 m / s, and repeat three times. Add 400 μL of DNA extraction buffer (10mM Tris HCL, pH7.5; 0.1mM EDTA, 6% SDS, 2% β-mercaptoethanol), mix well, and place in a 65°C water bath for 1 hour. Add 800μL of chloroformphenol (1:1), mix well, centrifuge at 12000g for 10min, and transfer the supernatant to a new centrifuge tube. Add 1 μL of glycogen (20 μg / μL), 40 μL...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com