ZmAGO1a protein as well as coding gene and application thereof
A technology that encodes genes and proteins, which can be used in applications, genetic engineering, plant genetic improvement, etc., and can solve problems such as lack of evidence at the molecular level.
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
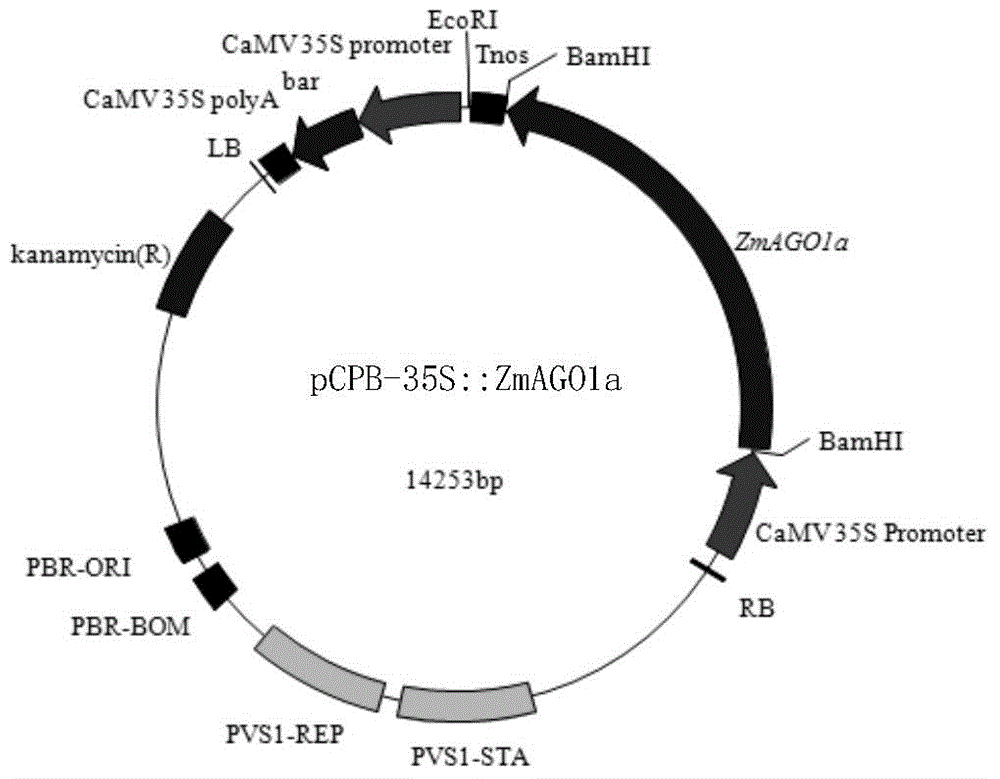
[0048] Example 1, Preparation of Plant Overexpression Vector pCPB-35S::ZmAGO1a
[0049] 1. The full-length cDNA sequence of the ZmAGO1a gene of the maize inbred line Ye 478 is shown in SEQ ID No. 1, and the 150th to 3458th positions in SEQ ID No. 1 are the coding gene sequence of the ZmAGO1a protein. The amino acid sequence of the ZmAGO1a protein is shown in SEQ ID No.2.
[0050] 2. Construction of pCPB-35S::ZmAGO1a
[0051] The sequence shown in the 123rd to the 3610th nucleotide in SEQ ID No.1 was inserted into the BamHI site of pCPB, and the remaining sequences of pCPB remained unchanged to obtain a recombinant plasmid, which was named pCPB-35S::ZmAGO1a, The pCPB-35S::ZmAGO1a was sent for sequencing, and the result was correct. The sequencing results showed that the sequence shown in the 123rd to 3610th nucleotides in SEQ ID No.1 was correctly inserted into the BamHI site of pCPB, and SEQ ID No. 2 shows the ZmAGO1a protein.
Embodiment 2
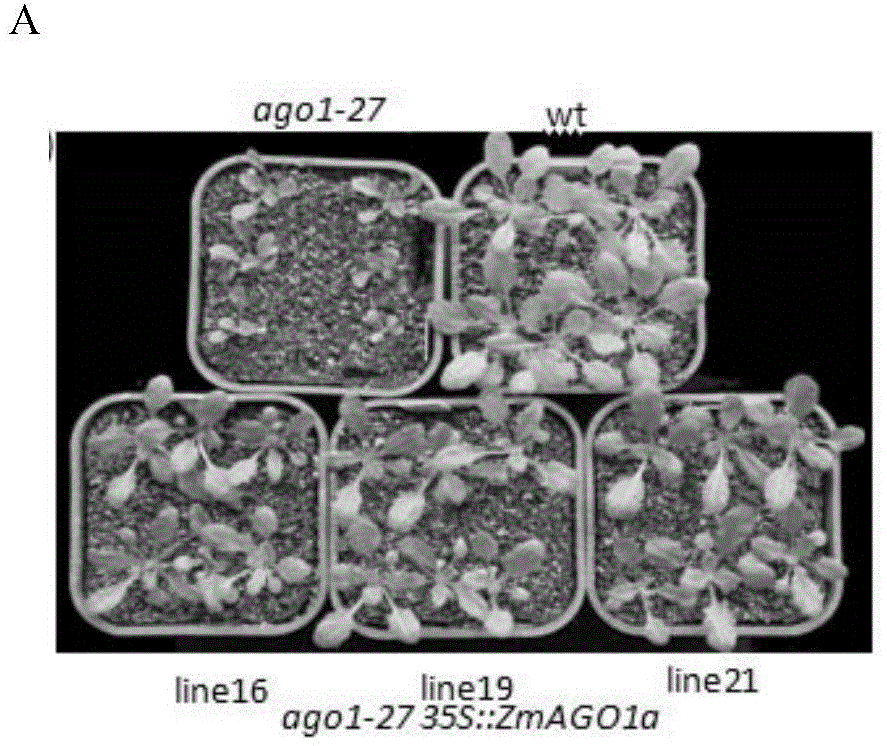
[0053] Example 2. Functional complementation identification of Arabidopsis ago1 mutant by ZmAGO1a gene
[0054] 1. After sterilizing the seeds of the Colombian ecotype Arabidopsis thaliana and the Arabidopsis ago1-27 mutant, they were placed in 1 / 2MS (Murashige and Skoog) medium respectively, under short-day light (8h light / 16h dark), 23 Cultivate at ±1°C for 10 days, then transfer to soil, and cultivate under short-day light (8h light / 16h dark) at 23±1°C.
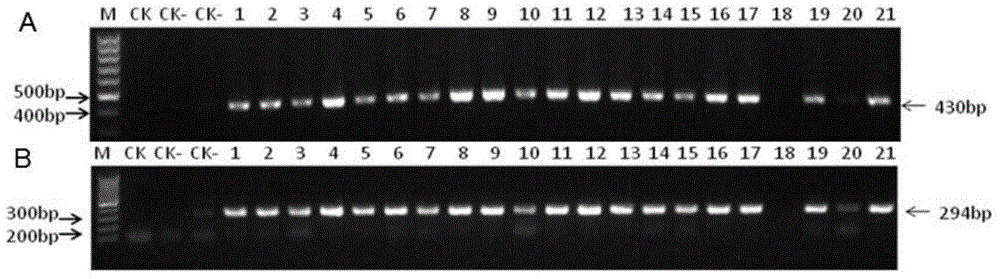
[0055] 2. Transform the plant overexpression vector pCPB-35S::ZmAGO1a prepared in Example 1 into Agrobacterium GV3301 to obtain recombinant Agrobacterium, and use the inflorescence infiltration method (Clough SJ, et al., 1998) to infect the recombinant Agrobacterium A cultured Colombian ecotype Arabidopsis thaliana and Arabidopsis ago1-27 mutant, respectively obtained Columbia ecotype Arabidopsis transgenic plants and Arabidopsis ago1-27 mutant transgenic plants, and harvested Columbia ecotype Arabidopsis transgenic plants...
Embodiment 3
[0099] Example 3, Tissue-specific analysis of ZmAGO1a gene expression in different organs of maize
[0100] 1. Collect the tassels at the tasseling stage, silk at the silking stage, bracts at the silking stage, and ear ears at the silking stage of the corn inbred line Ye 478 that grows normally in the field, and three of the normal hydroponic corn inbred line Ye 478. Leaf sheaths, young roots, and young leaves at the leaf stage.
[0101] 2. Extract the total RNA of each tissue collected in step 1, and reverse transcribe to obtain cDNA, respectively use the extracted cDNA as a template, and use ZmAGO1a forward primer (SEQ ID No.7) and ZmAGO1a reverse primer (SEQ ID No. .8) Perform real-time fluorescent quantitative PCR as primers to detect the relative expression level of the ZmAGO1a gene, or use the GAPDH forward primer and GAPDH reverse primer as primers to perform real-time fluorescent quantitative PCR to detect the relative expression level of the internal reference gene GA...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com