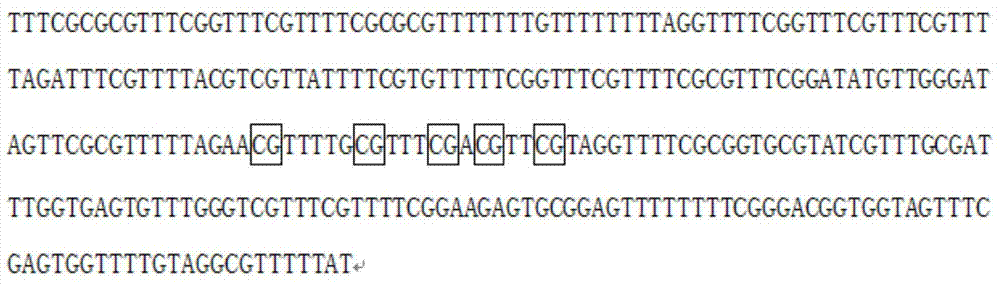Reagent and method for detecting MGMT gene promoter methylation
A technology for detection of reagents and promoters, applied in the fields of life science and biology, can solve problems such as variation of CG content and TM value, influence on acquisition, etc., and achieve high detection rate, high accuracy, and high throughput
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0045] The primers for detecting the methylation of the MGMT promoter include a pair of specific amplification primers SEQ NO 1 and SEQ NO 2, wherein the 5-end of SEQ NO2 is labeled with biotin, and the sequencing primer SEQ NO 3, its base sequence is as follows:
[0046] SEQ NO 1: 5'-GYGTTTYGGATATGTTGG-3';
[0047] SEQ NO 2: 5'-CRAAACRACCCAAACACTCAC-3';
[0048] SEQ NO 3: 5'-GATAGTTYGYGTTTTTAGAA-3';
[0049] When designing primers, it is considered that after the DNA sequence is treated with sulfite, some of its C bases will be converted into T, resulting in a large degree of variation in the CG content and TM value in the sequence region, which will affect the conventional primer design software in its sequence. Get the ideal primer sequence. Therefore, the amplification primers and sequencing primers used in PCR in the present invention are all self-designed, fully considering the characteristics of the DNA after sulfite treatment, and the binding site of the 3-terminal o...
Embodiment 2
[0060] Example 2: MGMT promoter methylation detection method
[0061] 1. The MGMT promoter methylation detection method is as follows:
[0062] (1) extract the DNA in the sample;
[0063] (2) subjecting the template DNA to sulfite treatment, and purifying and recovering;
[0064] (3) Using the purified recovered product in step (2) as a template, carry out two rounds of PCR amplification according to the specific amplification primers (SEQ NO1, SEQ NO2);
[0065] (4) After the PCR product is subjected to single-strand purification, it is placed in a pyrosequencer for sequencing;
[0066] (5) Obtain the methylation status of the MGMT promoter of the sample according to relevant software.
[0067] 2. Step (1) DNA extraction: You can choose extraction reagents well-known to those skilled in the art for DNA extraction, or use a kit developed by a biotechnology company for DNA extraction.
[0068] 3. Step (2) sulfite treatment and purification recovery methods are as follows: ...
Embodiment 3
[0079] Embodiment 3: clinical sample detection
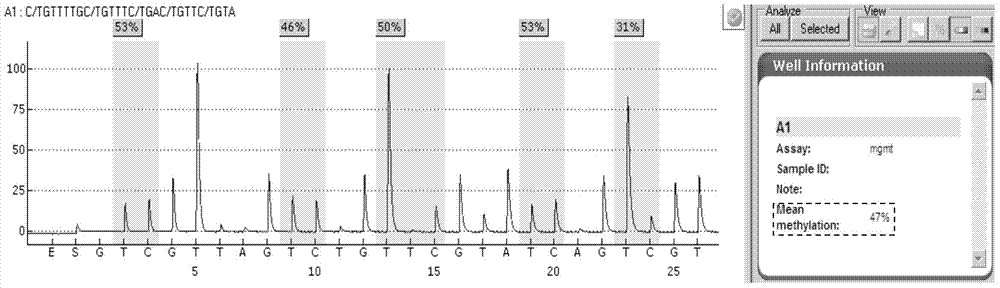
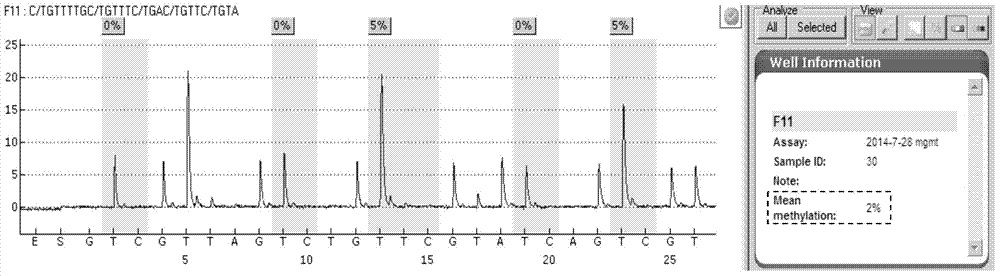
[0080] Select 36 cases of clinical tumor samples to be tested, and use the kit of the present invention for detection. Sample DNA extraction, sulfurous acid treatment, purification and recovery, PCR amplification, pyrosequencing and result analysis were carried out according to the method in Example 2. The results of the methylation status of the MGMT gene promoter of 36 samples are shown in Table 1:
[0081] Table 1
[0082]
[0083] The value in the column of methylation percentage of each sample in Table 1 represents the average methylation percentage value of the 5 detection sites on the MGMT promoter, which is given by the software PyroMark CpG after analysis.
[0084] It can be seen from Table 1 that the methylation status of the MGMT gene promoter in the above 36 tumor samples can be successfully and accurately detected using the detection method of PCR combined with pyrosequencing of the present invention. Therefore...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com