Kit and method for detecting methylation level of liver cancer risk gene TSPYL5
A methylation and kit technology is applied in the field of kits for detecting the methylation level of liver cancer risk gene TSPYL5, which can solve the problems of time-consuming, capital and energy-consuming, expensive and cumbersome process.
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
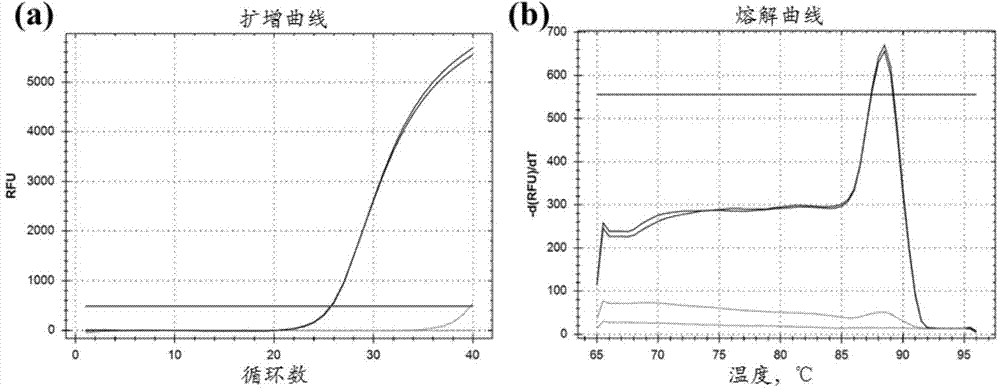
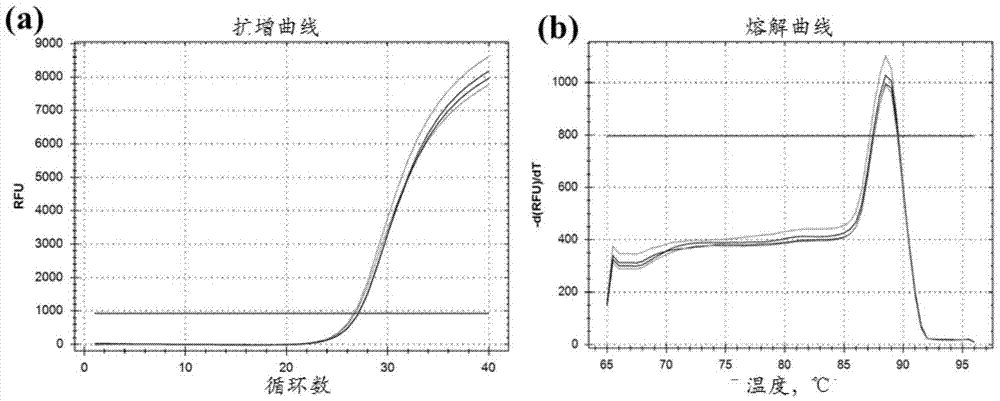
[0065] Example 1 TSPYL5 Gene methylation fluorescent PCR detection of 163 pairs of liver cancer and paracancerous tissue samples
[0066] Genomic DNA of paraffin samples and frozen tissue samples were extracted using kits from Qiagen (QIAamp DNA FFPE Tissue kit; QIAamp DNA Mini Kit). The concentration and purity of DNA samples were detected by NanoDrop-2000c.
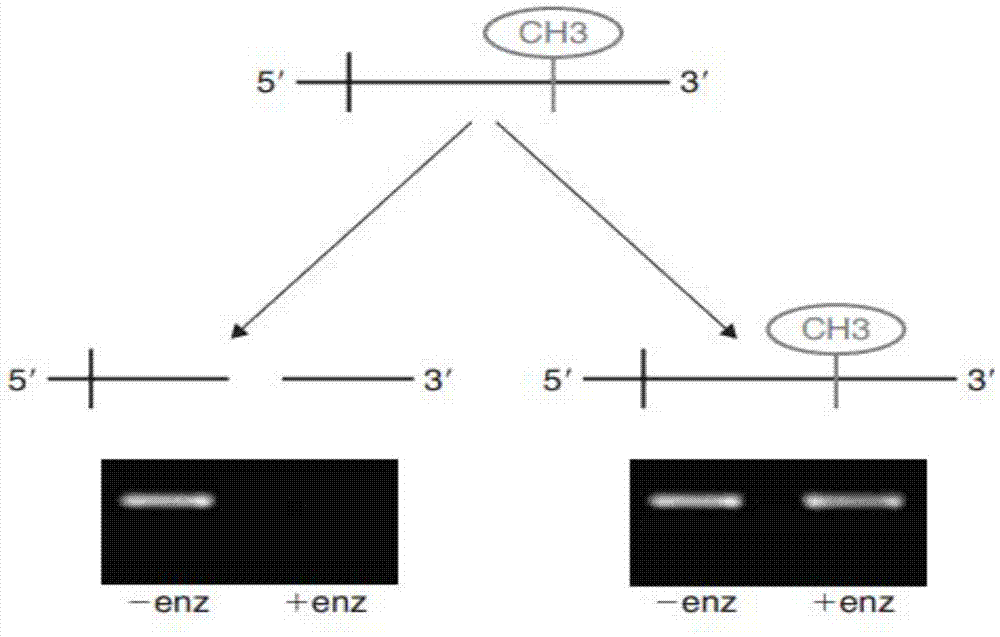
[0067] 300ng DNA sample was digested with methylation-sensitive restriction endonuclease (HinP1I). The digestion system was: DNA 300ng, Buffer (10×) 5μL, methylation-sensitive restriction endonuclease 30U, ddH 2 O supplemented to 50μL, this is the enzyme group. In addition to the treatment without enzyme group, ddH 2 Except for the restriction endonuclease replaced by O, the others are all the same as the enzyme addition group. Negative control (commercial unmethylated human genomic DNA control) and positive control (commercial fully methylated human genomic DNA) were also treated in the same way.
[0068] Design...
Embodiment 2
[0074] Example 2 BSP cloning sequencing method to detect liver cancer and paracancerous tissue TSPYL5 Gene methylation level
[0075] Genomic DNA of paraffin samples and frozen tissue samples were extracted using kits from Qiagen (QIAamp DNA FFPE Tissue kit; QIAamp DNA Mini Kit). The concentration and purity of DNA samples were detected by NanoDrop-2000c.
[0076] 2 μg of DNA samples were modified with sodium bisulfite, and the negative control (commercial unmethylated human genomic DNA control) and positive control (commercial fully methylated human genomic DNA) were also treated in the same way. The specific process is as follows :
[0077] (1) Take 2 μg of DNA and place it in a 2 mL EP tube, add appropriate amount of autoclaved water to make up the volume to 50 μL;
[0078] (2) Add 5.5 μL of freshly prepared 3mol / L NaOH, and bathe in water at 55°C for 20 minutes;
[0079] (3) Add 30 μL of freshly prepared hydroquinone solution (0.04 mol / L) and 520 μL of sodium bisulfi...
PUM
| Property | Measurement | Unit |
|---|---|---|
| Sensitivity | aaaaa | aaaaa |
| Sensitivity | aaaaa | aaaaa |
Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com