Reagent kit for detecting cancer-related DNA methylation and application of reagent kit for detecting cancer-related DNA methylation
A methylation and gene technology, applied in recombinant DNA technology, DNA/RNA fragment, determination/inspection of microorganisms, etc., can solve problems such as difficult diagnosis, and achieve the effect of good specificity and high diagnostic sensitivity
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0041] Embodiment 1, synthetic PFPN
[0042]Add 1,6-dibromohexane (23 mL, 154 mmol) and 2,7-dibromofluorene (5.01 g, 15.4 mmol) into a 150 mL one-necked bottle, heat and stir to dissolve. 50 mL of 50% KOH aqueous solution was added to the above reaction system, followed by tetrabutylammonium bromide (0.7 g, 2.17 mmol), and the reaction mixture was heated to 75° C. for 2 h. After the reaction was completed, the reaction mixture was extracted with dichloromethane (50 mL×2), the organic phases were combined, washed with water, 1M hydrochloric acid, washed with water, dried over anhydrous magnesium sulfate, and then the solvent was removed by rotary evaporation. Excess 1,6-dibromohexane was distilled off under reduced pressure to obtain a brown crude product. The crude product was separated and purified by silica gel column chromatography with petroleum ether as the eluent to obtain 2,7-dibromo-9,9-bis(6-bromohexyl)fluorene (7.40 g, 74%). 1 H NMR (300MHz, CDCl 3 ,ppm):7.53(d,2H...
Embodiment 2
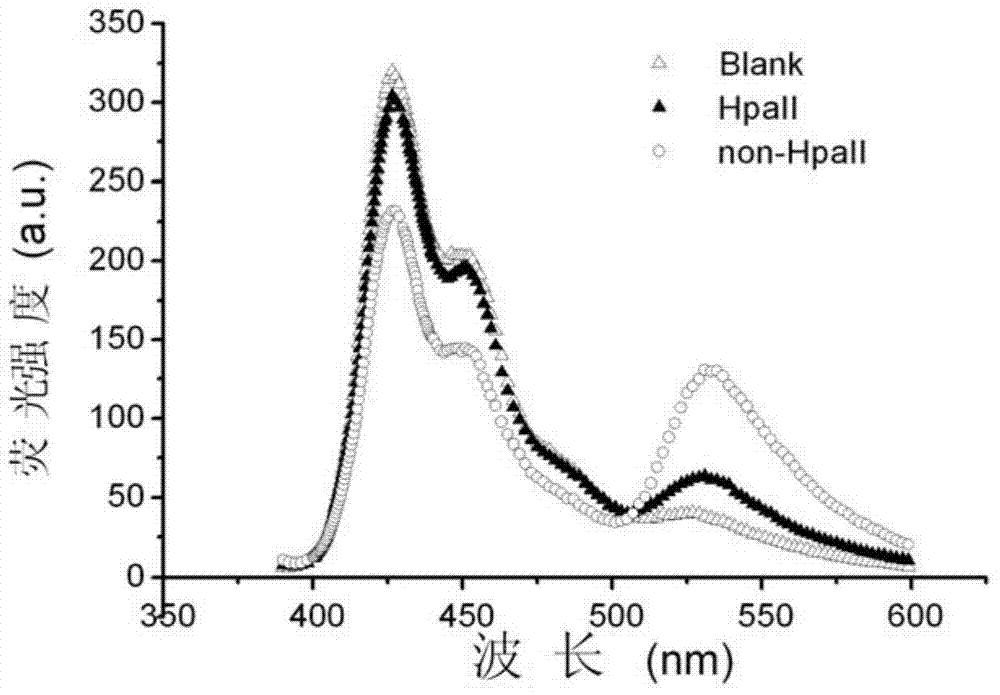
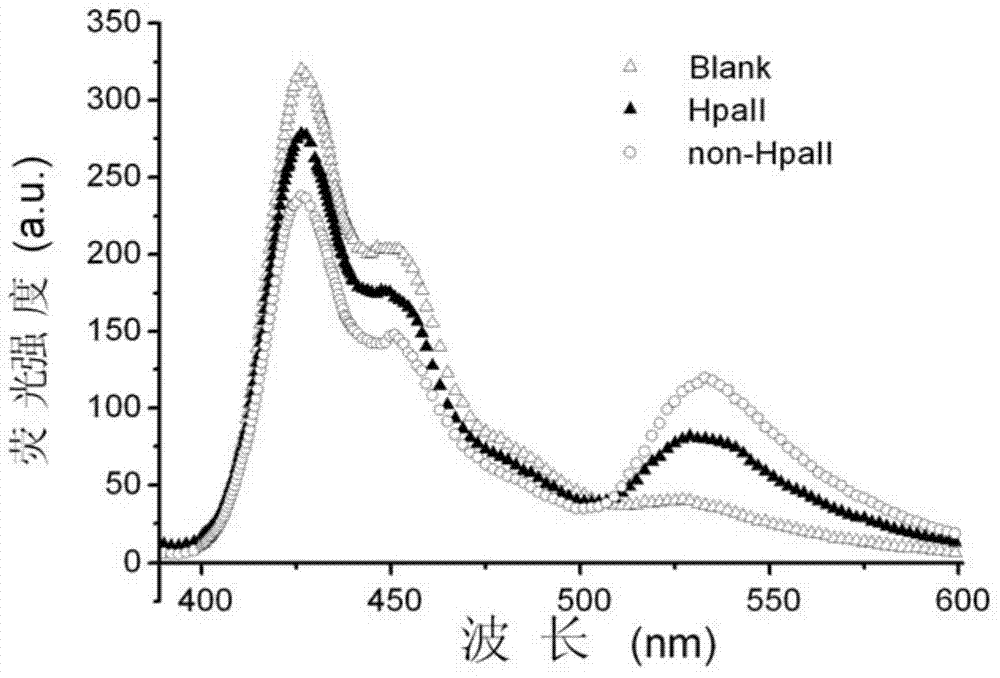
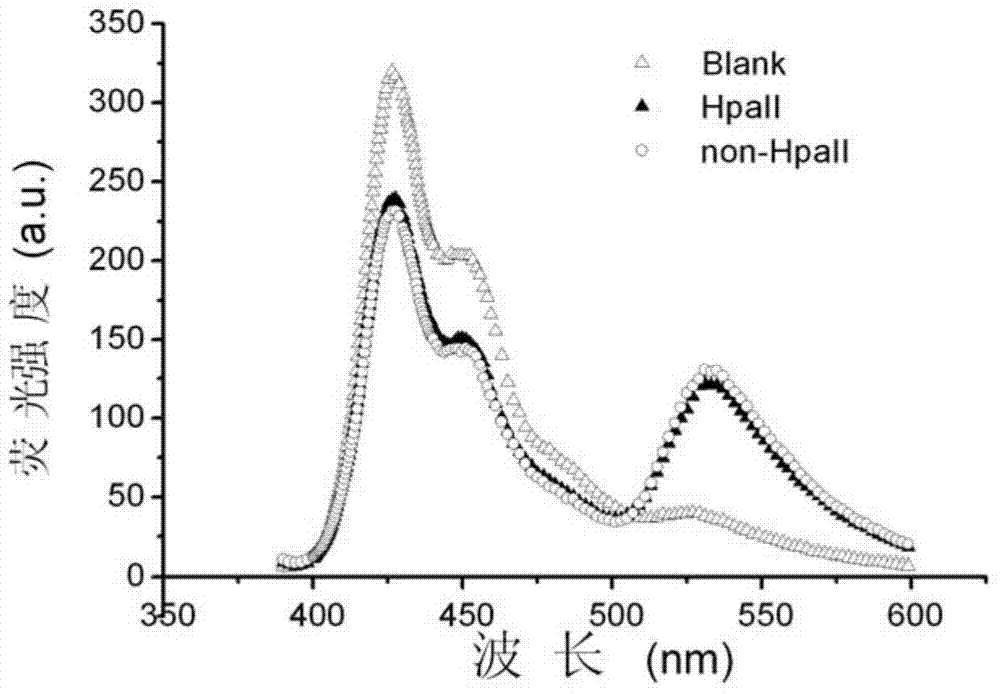
[0048] Example 2, PFPN-based FRET technology to detect DNA methylation to be tested
[0049] The following examples select genes whose promoter regions should be unmethylated in normal samples and methylated in cancer samples. According to literature reports, three related genes RASSF1A, OPCML and HOXA9 were selected in the experiment. All gene sequences can be found at http / / :www.ensembl.org. The designed amplified region (belonging to the promoter region) should contain 2 or 3 5'-CCGG-3' sites, where CG methylation is related to ovarian cancer and cannot be at the primer position.
[0050] The acquisition of genomic DNA
[0051] 35 ovarian cancer tissue samples and 11 normal tissue samples were selected from Changzhou Maternal and Child Health Hospital. All samples were obtained with informed consent. Genomic DNA was extracted from formalin-fixed and paraffin-embedded tissue samples with TIANamp gene extraction kit, and the genomic DNA of 35 ovarian cancer tissue samples...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com