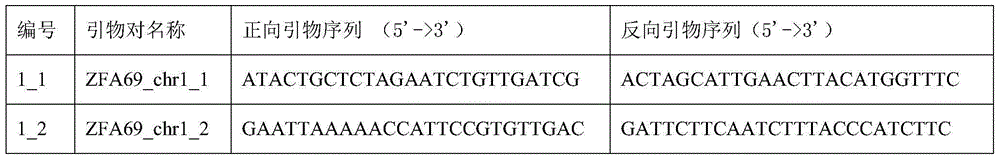Identification method for hybrid rice variety hybrid source superior 69 based on InDel (insertion-deletion length polymorphism) marker
An identification method and technology of hybrid rice, which is applied in the field of identification of hybrid rice variety Jiaoyuanyou 69, can solve the problems of not being able to obtain test results on the same day, expensive SNP chips, cumbersome data processing, etc., and achieve good application prospects and high accuracy , the effect of the simple method
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0027] Example 1. Map-based cloning of rice functional genes
[0028] Using the primer pair used in the invention and a mutant positioning population constructed based on a japonica-indica hybrid to carry out map-based cloning of rice genes.
[0029] The amplification system used is: 20μL, including 30-50ng template DNA, 0.25 units of DNA polymerase (Sheneng Group), 2.0μL of 10×Buffer (containing Mg2+), 2.0mM dNTPs 2μL, 1.0μL of two Methyl sulfoxide (DMSO);
[0030] The amplification program used was: 95°C pre-denaturation for 5 minutes; 95°C denaturation for 30s, 56°C renaturation for 30s, 72°C extension for 30s, 32 cycles; finally 72°C for 10min extension.
[0031] Electrophoresis detection: add an appropriate amount of loading buffer to the amplified product described in step 2, under 22V / cM constant electric field intensity, after 100 minutes of 10% denaturing polyacrylamide gel electrophoresis, silver staining develops; contains rice When the DNA sample is combined with the afo...
Embodiment 2
[0032] Example 2: Identification of germplasm resources of japonica rice varieties and indica rice varieties
[0033] The following primer pairs of the invention were used to identify the rice variety Jiaoyuanyou 69.
[0034]
[0035]
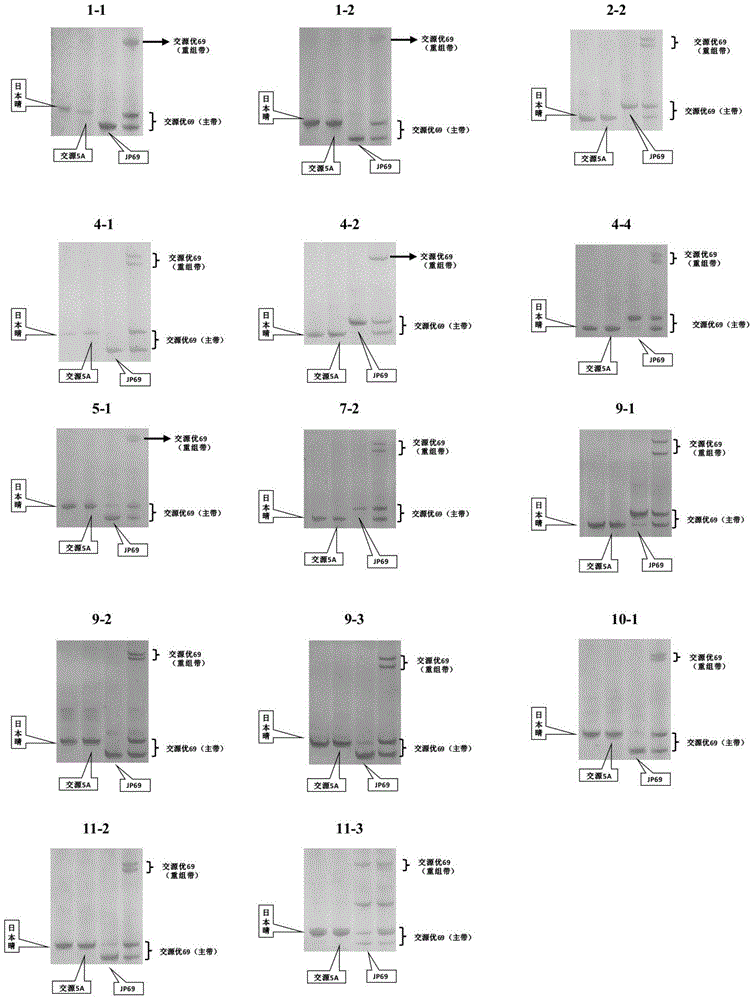
[0036] InDel molecular markers were used to identify rice varieties Nipponbare, Jiaoyuan 5A, JP69 and Jiaoyuanyou 69.
[0037] The amplification system used is: 20μL, including 30-50ng template DNA, 0.25 units of DNA polymerase (Shenergy Group), 2.0μL of 10×Buffer (containing Mg 2+ ), 2.0mM dNTPs 2μL, 1.0μL dimethyl sulfoxide (DMSO);
[0038] The amplification program used was: 95°C pre-denaturation for 5 minutes; 95°C denaturation for 30s, 56°C renaturation for 30s, 72°C extension for 30s, 32 cycles; finally 72°C for 10min extension.
[0039] Electrophoresis detection: add an appropriate amount of loading buffer to the amplified product described in step 2, under 22V / cM constant electric field intensity, after 100 minutes of 10% denaturing polyacrylam...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com