QPCR (quantitative polymerase chain reaction) primer for detecting NS2 genes of rice stripe viruses
A rice streak virus, target gene technology, applied in microorganism-based methods, DNA/RNA fragments, microorganisms, etc., can solve the problem of less protein research, and achieve the effect of reasonable primer dosage and guaranteed specificity
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
example 1
[0053] Example 1 Primer Specificity Detection
[0054] rice stripe virus Rice stripe virus , RSV), rice grass dwarf disease ( Ricegrass systunt virus , RGSV), rice sawtooth virus ( Riceragged stunt virus , RRSV) cDNA as a template, the primer sequence is NS2-qPCRF: ATCAACAGACGGAGCATC; NS2-qPCRR: CATTGGTTACAACTATGTGCTT. According to the following PCR system and procedures:
[0055] PCR reaction system:
[0056]
[0057] PCR reaction conditions:
[0058]
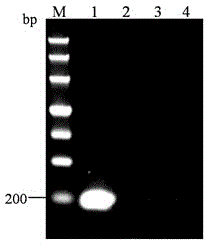
[0059] Judgment of results: After the reaction, judge the specificity of the primers according to the agarose electrophoresis graph. The electrophoresis graph shows that only the cDNA of RSV can amplify the band, and the band is single without primer-dimer, while the cDNA of RGSV and RRSV cannot. Amplified bands ( figure 1 ), which shows that the primer can specifically detect the gene of RSV NS2 .
example 2
[0061] Rice stripe virus ( Rice stripe virus , RSV), rice grass dwarf disease ( Ricegrass systunt virus , RGSV), rice sawtooth virus ( Riceragged stunt virus , RRSV) cDNA as a template, according to the following system and procedures (3 repetitions for each sample):
[0062] PCR reaction system:
[0063]
[0064] PCR reaction conditions:
[0065]
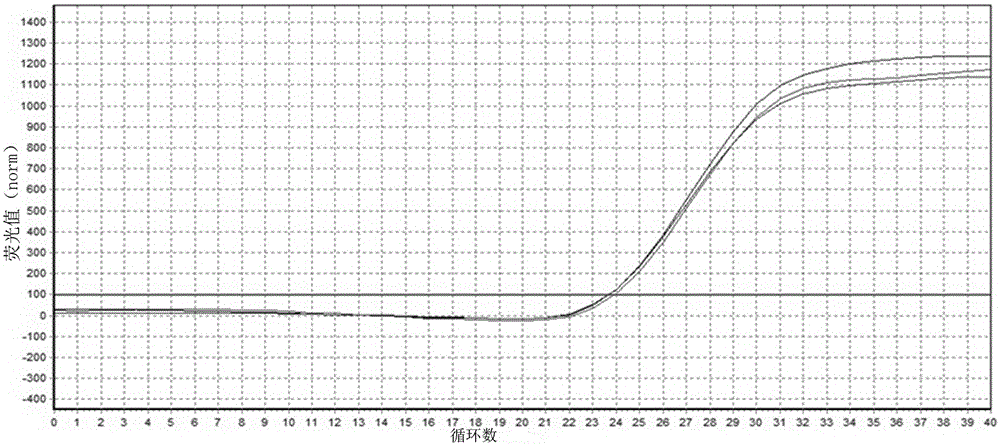
[0066] Result judgment: after the reaction finishes, judge the result according to the amplification curve diagram, and the RSV sample begins to peak at a Ct value of 20-25 and the peak value is high ( figure 2 ); while other samples only start peaking when the Ct value is higher than 35, and the peak value is low. Therefore, if the primer can peak before the Ct value is 30 ( image 3 ), it can be judged as the gene of RSV NS2 , otherwise, it cannot be judged.
example 3
[0068] Using the rice cDNA infected with RSV as a template, the rice elongation factor eEF1A (GenBank accession number: GQ848058) is an internal reference gene, which is carried out according to the following system and procedures (3 replicates for each sample):
[0069]
[0070] PCR reaction conditions:
[0071]
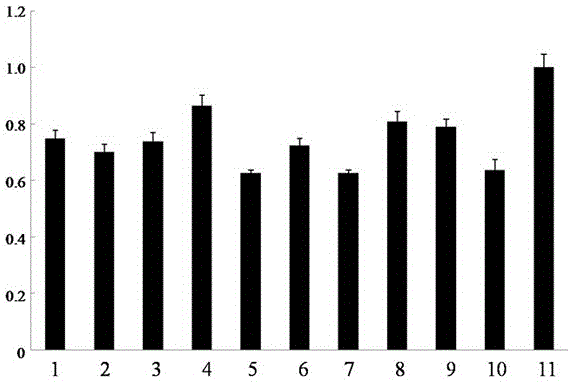
[0072] Judgment of results: After the reaction, according to the peak value of each sample and the qPCR calculation formula, the NS2 The expression level analysis graph of , which is a bar graph, and the height of the bar indicates the target gene in the RSV-infected rice sample NS2 The relative expression level can be judged by the graph NS2 The level of expression ( Figure 4 ), which shows that the primer can be used as a detection RSV NS2 qPCR primers for relative gene expression.
PUM
| Property | Measurement | Unit |
|---|---|---|
| PCR efficiency | aaaaa | aaaaa |
Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com