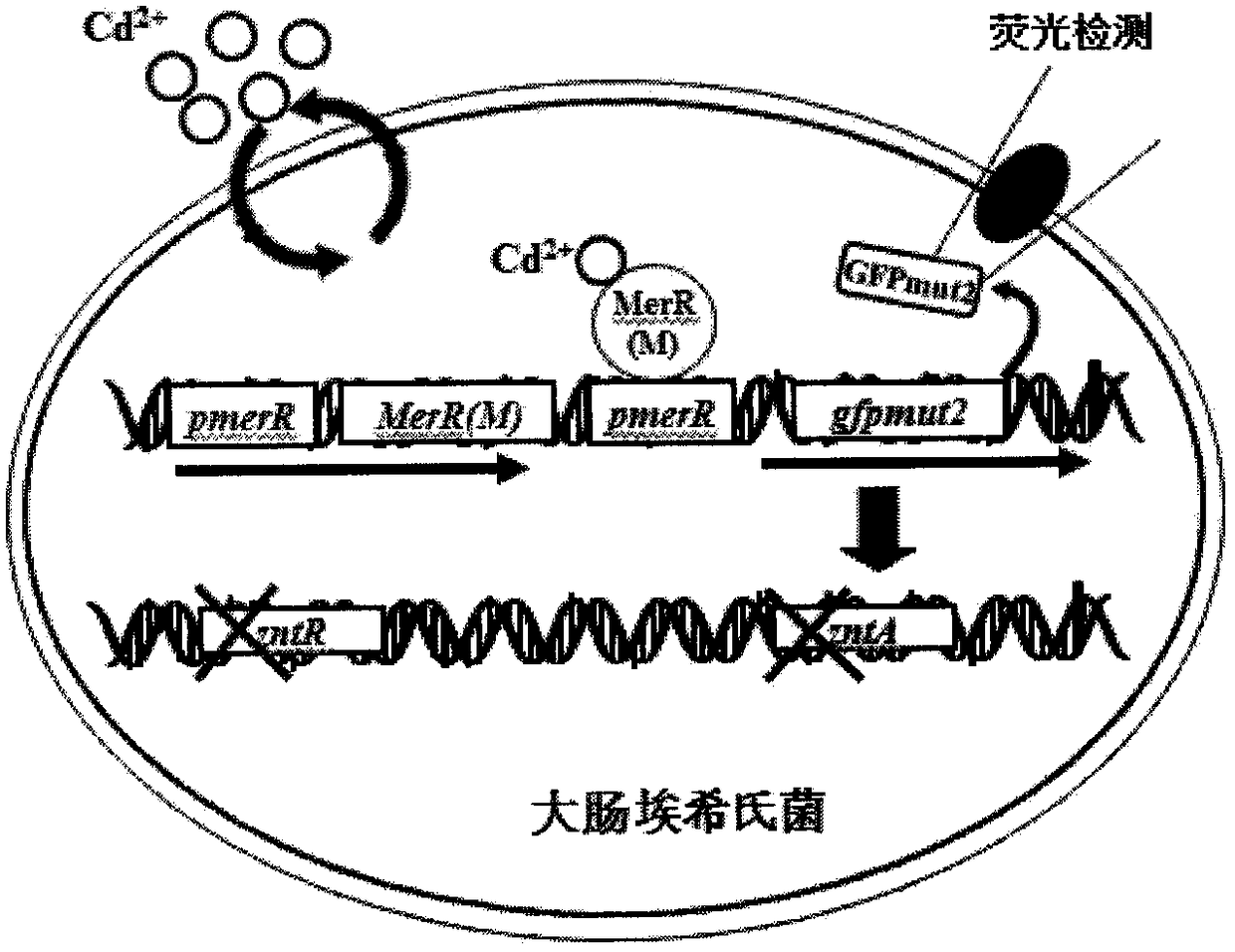
A strain of Escherichia coli detecting cadmium
A technology of Escherichia coli and escherichiacoli, which is applied in the direction of bacteria, microbe-based methods, and microbiological determination/testing, can solve the problems of no literature reports on engineering strains, and achieve reliable results, good repeatability, and detection signals stable effect
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
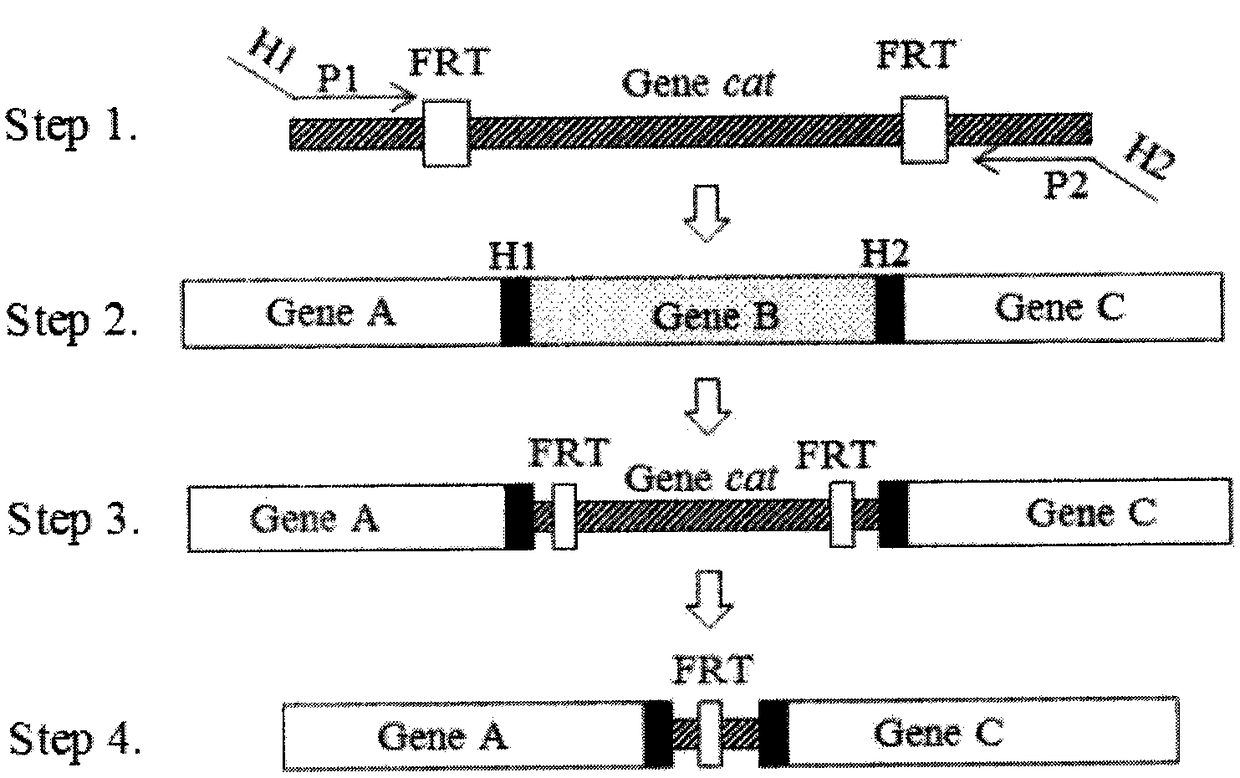
[0067] Embodiment 1. Preparation of mutant bacteria
[0068] 1. Primer information and synthesis
[0069] Gene knockout primers: According to the gene knockout primer sequence provided by http: / / ecogene.org / , Primer5.0 and DNAMAN software, design homologous recombination primers, see Table 1, the 5' end is the homology on both sides of the cat gene For the arm, see the ununderlined part in Table 1, and the 3' end is used to amplify the chloramphenicol resistance gene, see the underlined part in Table 1. Upstream homology arm primer zntA-Nf, sequence as shown in SEQ ID NO.6 and zntR-Nf, sequence as shown in SEQ ID NO.8; downstream homology arm primer zntA-Cr, sequence as shown in SEQ ID NO.7 Shown and zntR-Cr, the sequence is shown in SEQ ID NO.9.
[0070] Gene knockout identification primers: use a pair of primers spanning the gene to be knocked out provided by Harvard Molecular Technology Group & Lipper Center for Computational Genetics website http: / / arep.med.harvard.edu / l...
Embodiment 2
[0117] Embodiment 2. Construction of fusion reporter gene pmerR-MerR(M)-pmerR-gfpmut2
[0118] 1. Primer information and synthesis
[0119] According to the target gene pmerR and MerR(M) sequences published by GenBank and the known gfpmut2 gene sequence, use the Primer5.0 software to design primers, see Table 7, some primers introduce restriction sites at the 5' end or 3' end of the gene , design the primer Pmer-MerR-m-F for amplifying pmerR-MerR (M), the sequence is shown in SEQ ID NO.20 and Pmer-MerR-m-R, the sequence is shown in SEQ ID NO.21, amplify the pmerR-gfpmut2 gene The primer Pmer-GFPmut2-F sequence is shown in SEQ ID NO.22 and the GFPmut2-R sequence is shown in SEQ ID NO.23, and the final fusion fragment is connected to the identification primer M13F of the plasmid vector pMD19-T, and the sequence is shown in SEQ ID As shown in NO.24 and M13R, the sequence is shown in SEQ ID NO.25. See Table 7 for specific information, and the primers were synthesized by Invitrog...
Embodiment 3
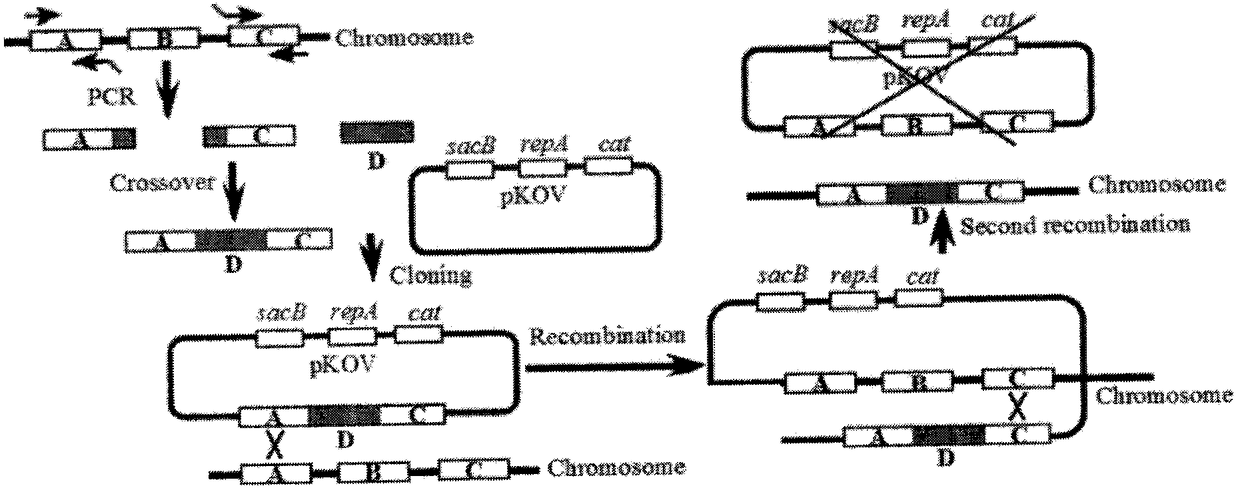
[0158] Example 3. Gene knock-in
[0159] 1. Primer information and synthesis
[0160] Use the Primer5.0 software to send out the primers, see Table 14, design the inner primers of the pmerR-MerR(M)-pmerR-gfpmut2 gene N-terminal and C-terminal, P 2 : zntR-cd-Ni, the sequence is shown in SEQ ID NO.15 and P 5 : zntR-cd-gfpmut2-Ci, the sequence is shown in SEQ ID NO.16 and the outer primer, P 1 : zntR-No, the sequence is shown in SEQ ID NO.14 and P 6 : zntR-Co, the sequence is shown in SEQ ID NO.17, making P 2 : zntR-cd-Ni with P 3 : pmerR-MerR(M)-pmerR-gfpmut2-F, the sequence is shown in SEQ ID NO.18, P 4 : pmerR-MerR(M)-pmerR-gfpmut2-R, the sequence is shown in SEQID NO.19, and P 5 : There is a complementary sequence between zntR-cd-gfpmut2-Ci, the gene knockout two outer primers remain unchanged, P 3 with P 4 A pair of gene primers for amplifying pmerR-MerR(M)-pmerR-gfpmut2, P 7 : pkov-SalI, sequence as shown in SEQ ID NO.28 and P 8 : pkov-NotI, the sequence is as sho...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com