A method for establishing a receptor protein model for n-glycosylation efficiency detection in Escherichia coli using the skeleton protein fn3
A technology of Escherichia coli and skeleton protein, applied in the direction of microorganism-based methods, chemical instruments and methods, biochemical equipment and methods, etc., can solve problems such as poor flexibility, achieve improved physical and chemical properties, high-efficiency and low-cost large-scale production, elucidation The effect of structure-activity relationship
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
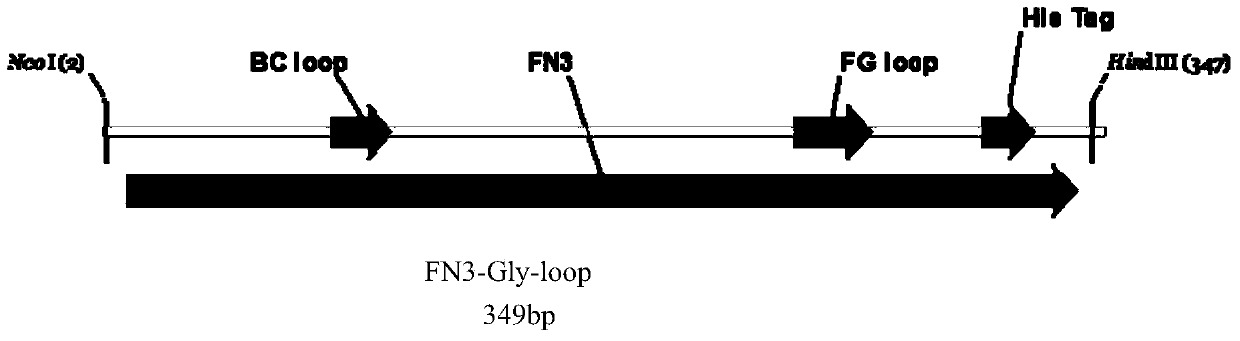
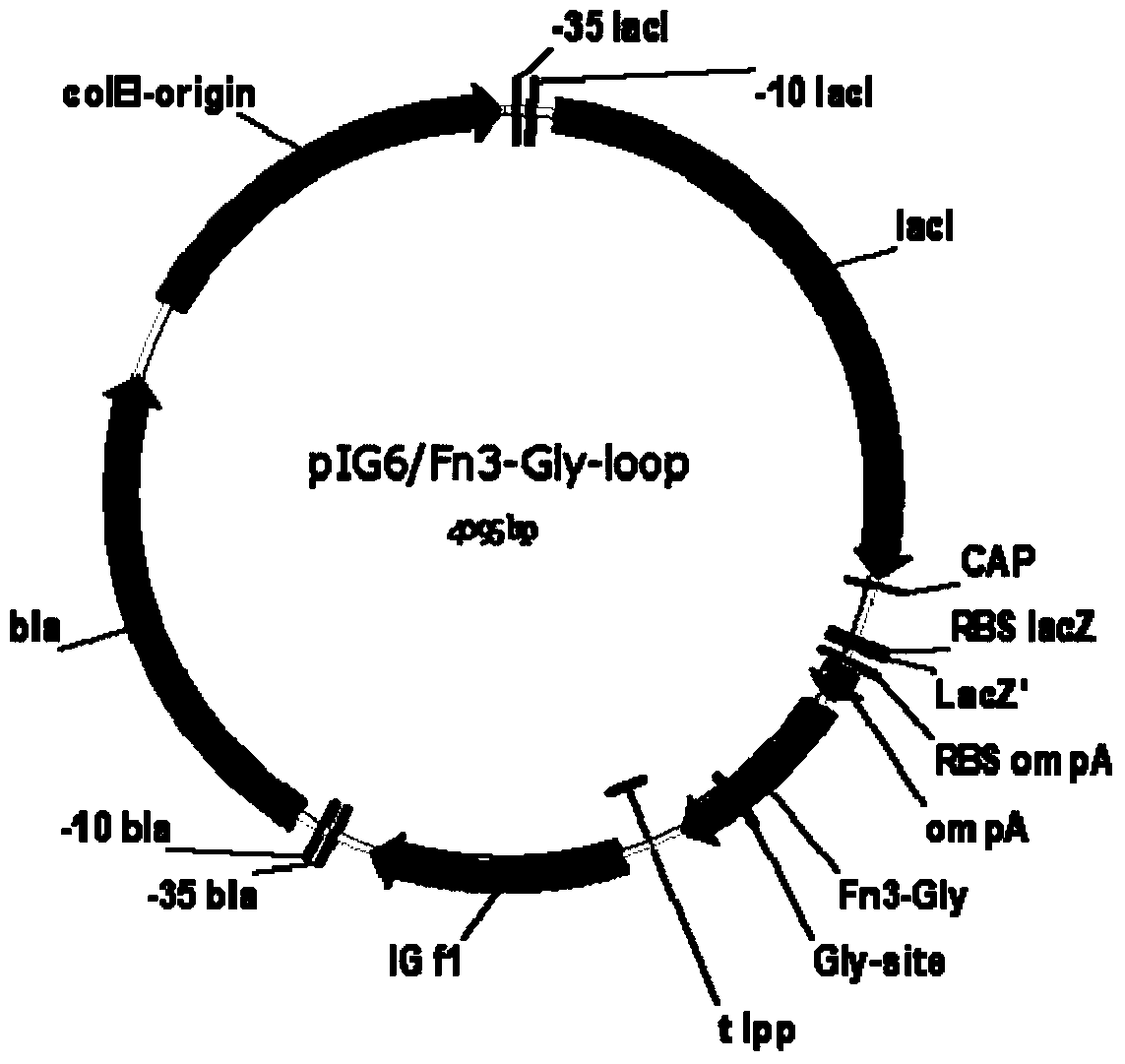
[0018] 1. In this example, human fibronectin type III domain (Fn3) protein was used as a model gene (EMBL accession number AJ320527). At the 5′ end of the gene, 6 histidine residues are introduced, and at the FGloop region, a base sequence encoding the glycosylation site DQNAT is introduced. See the structure of the fusion gene. figure 1 . The gene was constructed into pIG6H by gene recombination, see the vector structure figure 2 .
[0019] 2. After the human fibronectin type III domain mutant Fn3-Gly-loop gene expression cassette sequence shown in the sequence table was synthesized (Nanjing GenScript Biotechnology Co., Ltd.), EcoRV and HindIII were used to construct an E. coli expression vector On pIG6H, the recombinant vector pIG6H-Fn3-Gly-loop was obtained.
[0020] 3. the expression vector constructed and pACYCpgl electric shock transformation CLM37 escherichia coli bacterial strain, then transformants were inoculated to the LB solid medium (per liter of culture) cont...
Embodiment 2
[0024] 1. In this example, human fibronectin type III domain (Fn3) protein was used as a model gene (EMBL accession number AJ320527). Introduce 6 histidine residues encoding at the 5′ end of the gene, and introduce the nucleotide sequence encoding the glycosylation site DQNAT in the FG loop region. The structure of the fusion gene is shown in figure 1 . The gene was constructed into pIG6H by gene recombination, see the vector structure figure 2 .
[0025] 2. After the human fibronectin type III domain mutant Fn3-Gly-loop gene expression cassette sequence shown in the sequence table was synthesized (Nanjing GenScript Biotechnology Co., Ltd.), EcoRV and HindIII were used to construct an E. coli expression vector On pIG6H, the recombinant vector pIG6H-Fn3-Gly-loop was obtained.
[0026] 3. the expression vector constructed and pACYCpgl electric shock transformation CLM37 escherichia coli bacterial strain, then transformants were inoculated to the LB solid medium (per liter of...
Embodiment 3
[0030] 1. In this example, human fibronectin type III domain (Fn3) protein was used as a model gene (EMBL accession number AJ320527). At the 5′ end of the gene, 6 histidine residues are introduced, and at the FGloop region, a base sequence encoding the glycosylation site DQNAT is introduced. See the structure of the fusion gene. figure 1 . The gene was constructed into pIG6H by gene recombination, see the vector structure figure 2 .
[0031] 2. After the human fibronectin type III domain mutant Fn3-Gly-loop gene expression cassette sequence shown in the sequence table was synthesized (Nanjing GenScript Biotechnology Co., Ltd.), EcoRV and HindIII were used to construct an E. coli expression vector On pIG6H, the recombinant vector pIG6H-Fn3-Gly-loop was obtained.
[0032] 3. the expression vector constructed and pACYCpgl electric shock transformation CLM37 escherichia coli bacterial strain, then transformants were inoculated to the LB solid medium (per liter of culture) cont...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com