DNA-protein conjugate and application thereof in detecting mercury ion concentration
A conjugate and protein technology, applied in the field of detection and analysis, can solve problems such as difficult to reuse, high detection cost, and large consumption of reagents
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
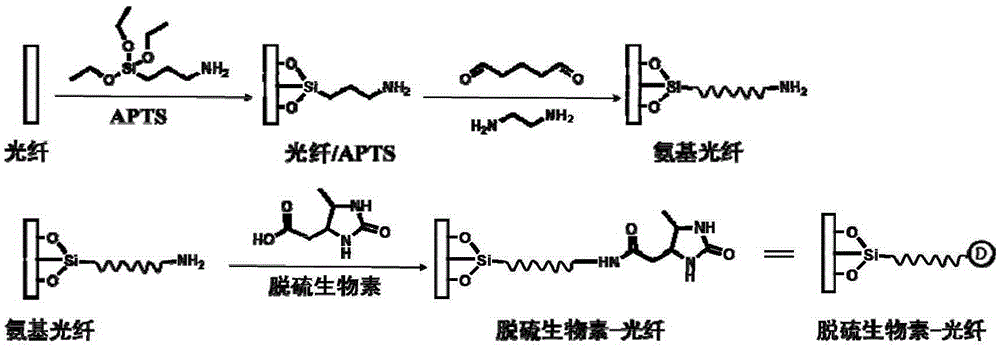
[0039] Example 1 Preparation of Dethiobiotin Modified Optical Fiber
[0040] 1. Optical fiber pretreatment
[0041] Put the optical fiber into the piraha solution (concentrated H 2 SO 4 :H 2 o 2(v / v)=3:1) in a colorimetric tube, heated at 120°C for 1 hour, after heating, fully washed the optical fiber with ultrapure water for more than 10 times, dried the optical fiber with nitrogen at room temperature, and placed it in a 70 °C overnight in a vacuum oven.
[0042] 2. Preparation of APTS-optical fiber
[0043] The next day, prepare 2% (volume ratio) (3-aminopropyl) triethoxysilane (APTS) solution with anhydrous toluene, and put the clean optical fiber into the solution for one hour reaction; rinse with toluene for 5 times, dry the optical fiber with nitrogen, and bake it at 200°C for 1 hour.
[0044] 3. Preparation of amino-fiber
[0045] After natural cooling, the optical fiber was immersed in 2.5% glutaraldehyde in ethanol solution, reacted at room temperature for 1 h...
Embodiment 2D
[0051] Synthesis of Example 2DNA-Protein Conjugates
[0052] 1. Preparation of activated thiol DNA
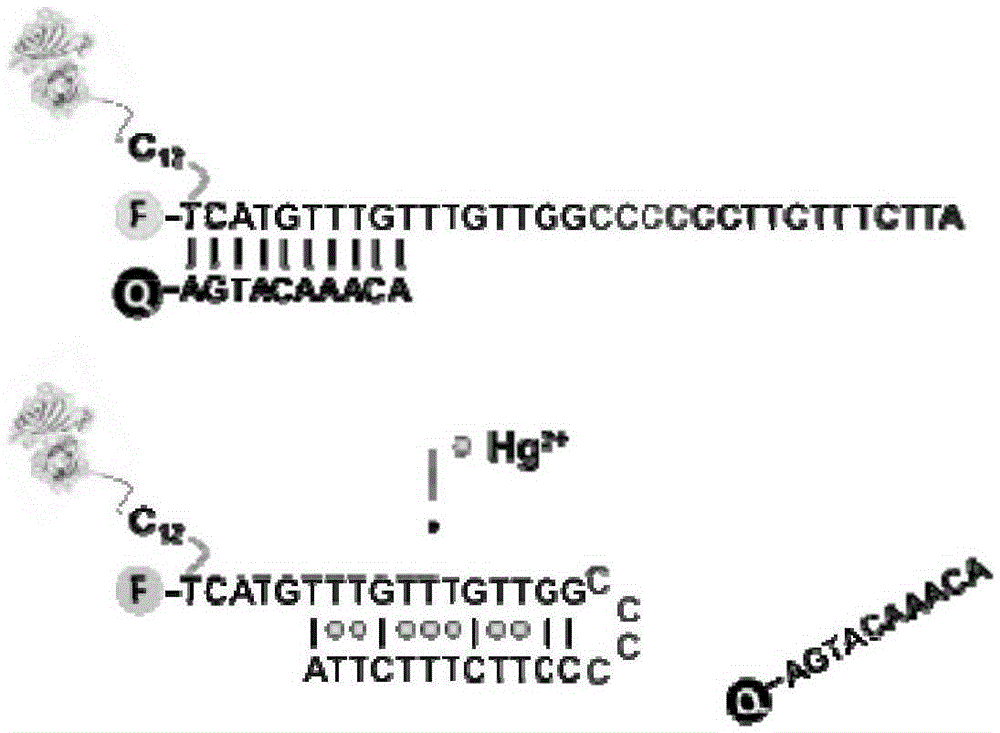
[0053] In 10 μl 1mMSH-DNA-Cy5.5 (specific DNA segments such as figure 1 Add 5 μL of 1M sodium phosphate buffer solution (pH=5.5) and 1 μL of 50 mMTCEP solution to the indicated) and place the mixture in the dark for 1 hour to react at room temperature; then add 200 μL of phosphate buffer solution (0.1M sodium phosphate, 0.15M sodium chloride , pH7.2, the same below), centrifuged at 12000rpm for 10 minutes, and repeated 10 times to obtain activated sulfhydryl DNA.
[0054] 2. Preparation of activated streptavidin protein
[0055] Add 0.5 mg Sulfo-SMCC to 150 μL 20 mg / mL streptavidin, vortex for 5 minutes, and let it react at room temperature for 1 hour. Use Amicon-50k to remove undissolved Sulfo-SMCC, add 200 μL phosphate buffer each time, and set the temperature at 12000 rpm Centrifuge for 10 minutes and repeat 10 times to obtain activated streptavidin protein.
[0056] 3. ...
Embodiment 3
[0060] The detection of mercury ion in embodiment 3 water bodies
[0061] 1. Configuration of mercury ion stock solution
[0062] Prepare mercury ion concentration gradient stock solutions with concentrations of 0.01 μg / mL, 0.1 μg / mL, 1 μg / mL and 10 μg / mL, respectively, for future use.
[0063] 2. Detection of mercury ions in water
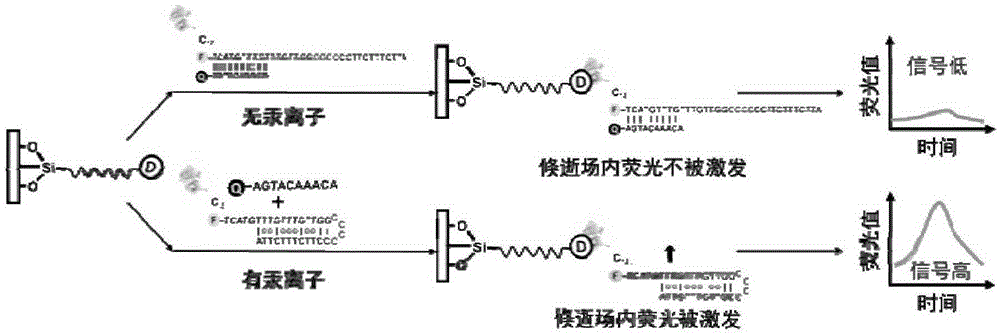
[0064] With MOPS buffer (10mM MOPS, 0.5MNaNO 3 , pH7.5) as the reaction solution. For a single detection, 2 μL of DNA-streptavidin protein conjugate stock solution and different concentrations of mercury ion gradient stock solutions were added to 450 μL of reaction solution, and the reaction was shaken at room temperature for 1 hour. The reacted liquid is passed into the evanescent wave optical fiber sensing platform.
[0065] During the optical fiber detection process, the optical fiber is first washed with MOPS buffer until the baseline is stable; after the baseline is stable, the liquid reacted with mercury ions is introduced, the peristalt...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com