Composite tag for high-throughput sequencing of biological diversity of fungi in environment and application of composite tag
A biodiversity and compound labeling technology, applied in the field of ultra-high-throughput gene sequencing and compound labeling, can solve the problems of great impact and disadvantages on the quality of wine
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0036] The extraction of total DNA from environmental samples can refer to Zhou's method (DNA recovery from soils of diverse composition, Appl. Environ. Microbiol., 1996, 62, 316-322), and the purification of extracted DNA can refer to Jackson's method (Asimple, efficient method for the separation of humic substances and DNA from environmental samples, Appl. Environ. Microbiol ., 1997, 63, 4993-4995). Or refer to the method of soil DNA extraction kit to extract. The main steps are as follows:
[0037] 1. Weigh 5 g of environmental samples into a 50 mL centrifuge tube, add 15 mL of phosphate buffer, vortex vigorously for 5 min on a vortexer, then centrifuge at 12,500 rpm for 10 min, discard the supernatant, and grind the precipitate with sterilized quartz sand;
[0038] 2. Add 13.5 ml of DNA extraction buffer, 100 microliters of 20 mg / mL proteinase K, shake at 37°C, 250 rpm for 30 minutes;
[0039] 3. Add 700 microliters of 20% SDS, water bath at 65°C for 2 hours, invert and sh...
Embodiment 2
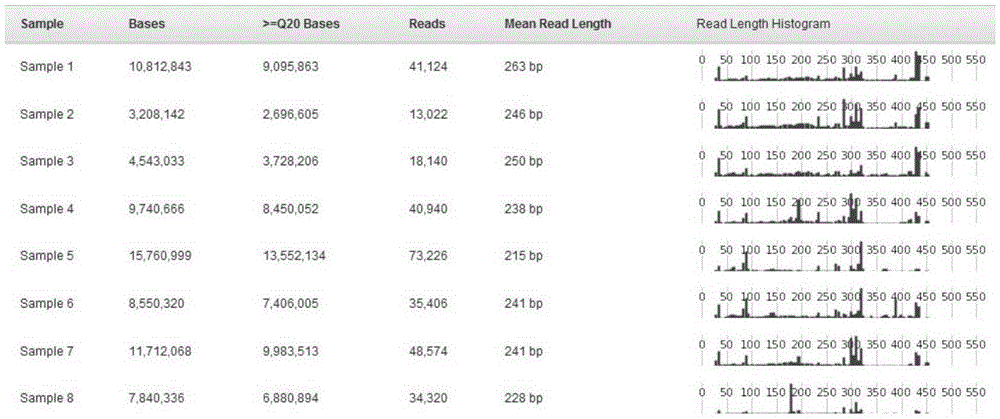
[0047] Take 8 samples as an example to implement.
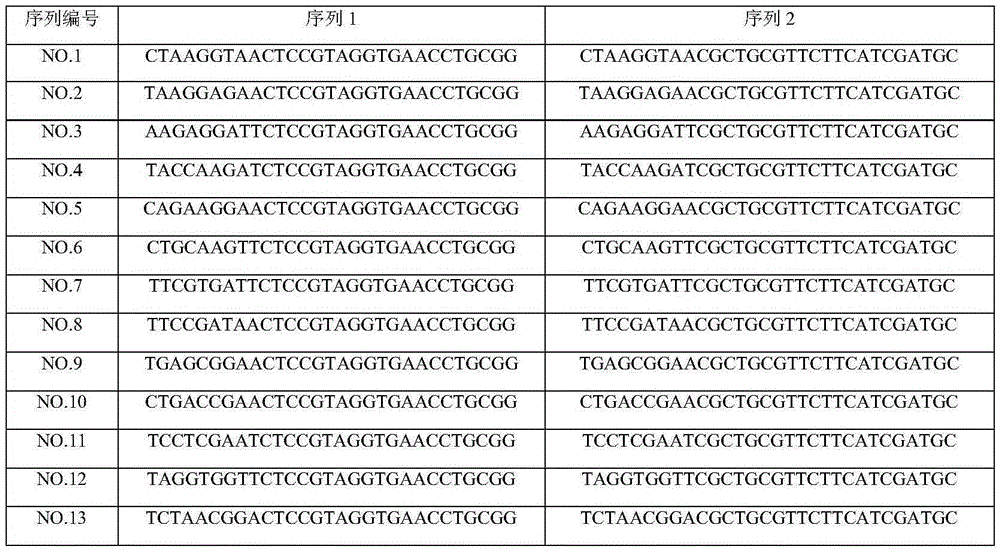
[0048] (1) Number the 8 samples from 1 to 8, and select a sequence number (as shown in Table 2) from the sequences No. 1 to No. 48 in Table 1 for synthesis; Dilute the synthesized sequences to 10 pmol with water; associate the sequence numbers in Table 1 with the corresponding sample numbers, as shown in Table 2.
[0049] Table 2 Correspondence table between samples and compound labels
[0050] sample number 1 2 3 4 5 6 7 8 serial number No.1 No.2 No.3 No.4 No.5 No.6 No.7 No.8
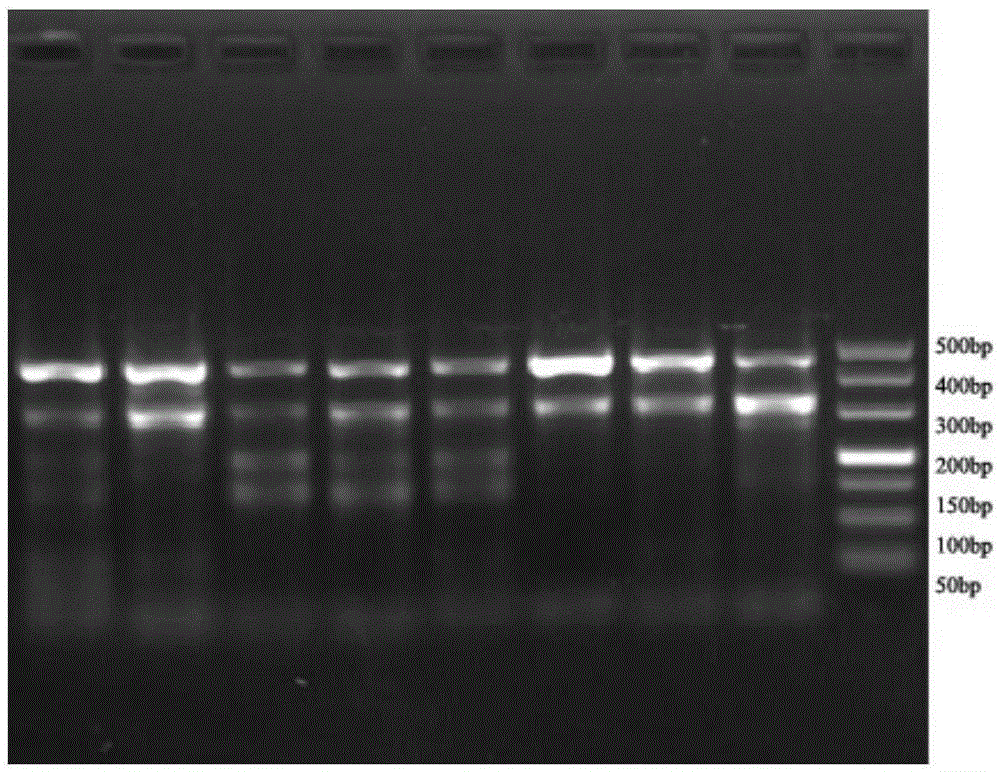
[0051] (2) PCR amplification: Take 8 200 μL PCR tubes, each tube corresponds to a sample, and use sequence 1 and sequence 2 in the sequence number corresponding to the above sample number as primers for PCR amplification. The PCR-amplified 50 μL system contains: according to the DNA concentrations of the 8 samples, draw a total of 50 ng of the total DNA of the samples into the corresponding tube, and then a...
Embodiment 3
[0055] 1. Use a general PCR product recovery kit to purify the PCR amplification product, the steps are as follows:
[0056] (1) Add 2 volumes of BindingBuffer to 1 volume of PCR reaction, invert and mix well.
[0057] (2) Add the above mixture to the DNA purification column. If the solution volume is >700 μl, transfer the solution in stages; leave it at room temperature for 1-2 min or longer.
[0058] (3) Centrifuge at 13000g for 1 minute. After the centrifugation, transfer the solution in the collection tube to the DNA purification column again, centrifuge, and discard the waste liquid.
[0059] (4) Add 650 μL WashBuffer to the DNA purification column, centrifuge at 13,000g for 30 seconds, discard the waste liquid, and put the DNA purification column back into the collection tube. Repeat step 4.
[0060] (5) Centrifuge at 13,000 g for 3 min to remove residual ethanol in the column.
[0061] (6) Put the purification column into a new centrifuge tube, add 30-50 μL of Elutio...
PUM
| Property | Measurement | Unit |
|---|---|---|
| Molecular weight | aaaaa | aaaaa |
Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com