Dual-ring hairpin probe mediate label-free strand displacement amplification method for detecting bleomycin
A technology of strand displacement amplification and hairpin probe, which is applied in the field of label-free strand displacement amplification mediated by double-loop hairpin probes for the detection of bleomycin, which can solve the problems of high background signal, false positive signal, etc.
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment Construction
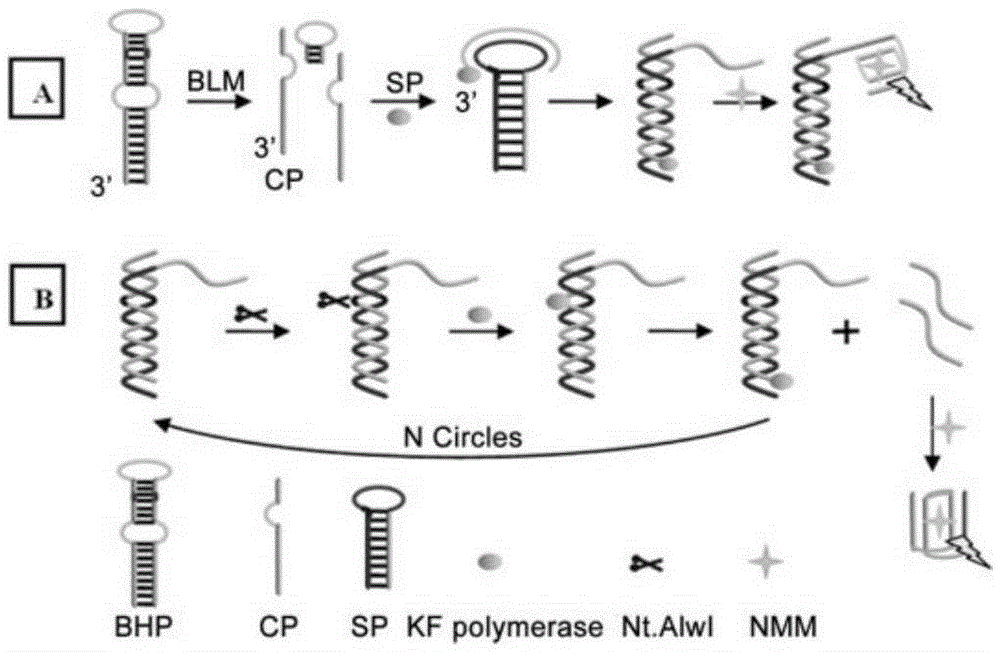
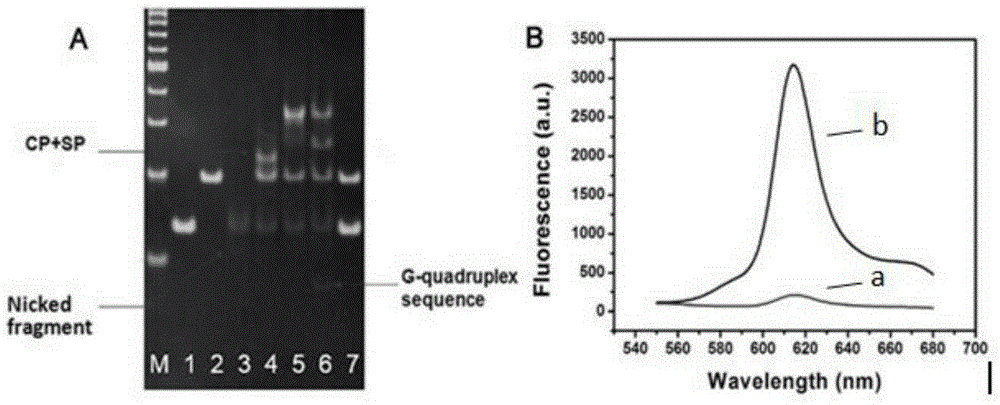
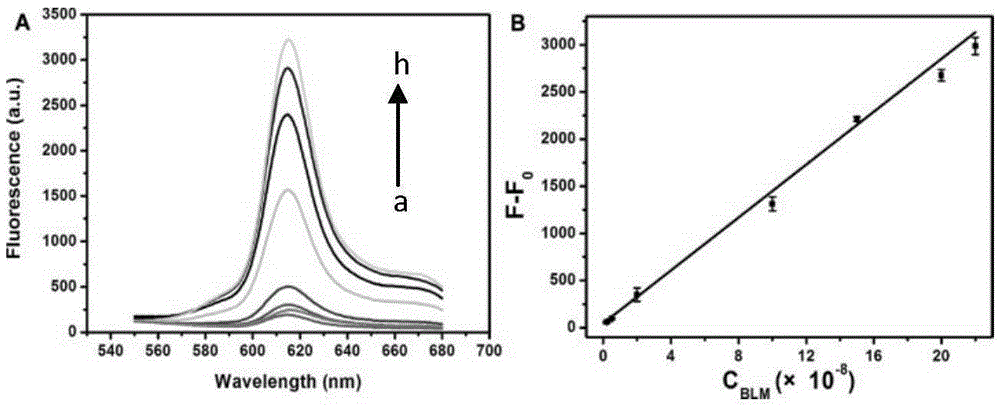
[0022] The present invention will be further described below with reference to the accompanying drawings and embodiments.
[0023] Materials and Instrumentation Bicyclic hairpin probe (BHP) and signal probe (SP) oligonucleotide sequences were synthesized and purified by Sangon Biotechnology Co., Ltd. (Shanghai, China). Bleomycin A5, mitomycin and daunorubicin were purchased from China National Institute for Food and Drug Control. Actinomycin D was purchased from Melone Pharmaceutical Co., Ltd. (Dalian, China). Ferrous chloride (FeCl 2 ● H 2 O) was purchased from Tianjin Guangfu Institute of Fine Chemicals. Klenowfragment (3'-5'exo-) polymerase and Nt.AlwI were purchased from NewEngland Biolabs Ltd. (Beijing, China). dNTPs were purchased from ThermoFisher Scientific Ltd. (China). NMM was purchased from Frontier Scientific Inc. (Utah, USA). Clinical serum samples were obtained from the Affiliated Hospital of Shandong University. All other chemical reagents are of analyti...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com