Fluorescent quantitative PCR (polymerase chain reaction) detection primers and kit for Eimeria media-rabbit
A technology for fluorescent quantitative detection and Eimeria coccidiosis, which is applied in the field of detection, can solve the problems of increasing the difficulty of differential diagnosis for clinical operators, and the morphological characteristics of oocysts are not very obvious, and achieve objective result judgment, high sensitivity, Simple operation effect
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0024] Example 1 Primer Design and Screening
[0025] Firstly, the ribosomal ITS sequence of Eimeria mesococcidia isolated from Yulin, Guangxi was amplified by PCR and sequenced, and sequence analysis was performed with the sequence published in GenBank. It was found that the ITS sequence showed relative conservation within the species and The characteristics of large differences among species are suitable as molecular marker sequences for molecular identification between species. Design specific primers in the conserved regions within the species, and refer to the specific primers reported abroad, and screen out the most suitable primers through experimental verification for quantitative experiments.
[0026] Table 1 Design of primers for quantitative PCR of rabbit Eimeria mesodium
[0027]
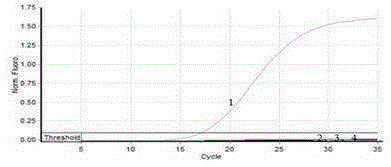
[0028] Fluorescent quantitative PCR was performed on the sequences designed in Table 1, and the results showed that only primer set 1 could specifically amplify.
[0029] In order t...
Embodiment 2
[0030] The preparation and use method of embodiment 2 kit
[0031] The kit contains stool DNA extraction reagent (QIAampDNAStoolMiniKit), which contains 100 reactions of 140mL ASLBuffer, 1.5mL proteinaseK, 250mL BufferAL, 50mL BufferAW1, 50mL BufferAW2, 10mL BufferAE, 20mL 96%-100% Alcohol; the fluorescent quantitative PCR reaction solution is 100 reactions of SYBRGreenPrimixExTaqII reagent, and the final concentration is 25 pmol / μL of Eimeria medusia specific primers.
[0032] One, the use method of above-mentioned kit, comprises the following steps:
[0033] (1) Extraction of stool DNA:
[0034] Refer to the instructions of QIAampDNAStoolMiniKit, the specific operation steps are as follows:
[0035] a. Weigh 200mg of feces and place it in a 2mL centrifuge tube.
[0036] b. Add 1.4mLASLBuffer and 0.4g glass beads (1mm), shake for 1-2h.
[0037] c. Place the suspension at 70°C for 5min.
[0038] d. Vortex for 15 seconds, then centrifuge the sample at 15,000 rpm for 1 minute...
Embodiment 3
[0062] The sensitivity test of embodiment 3 kits
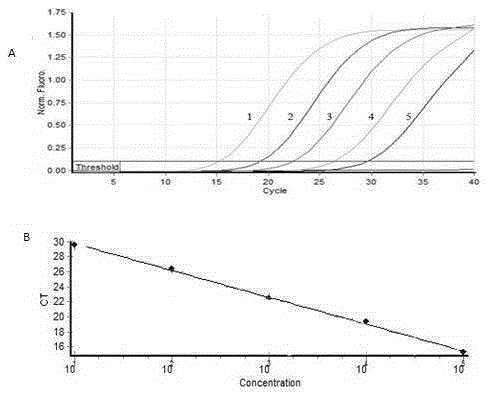
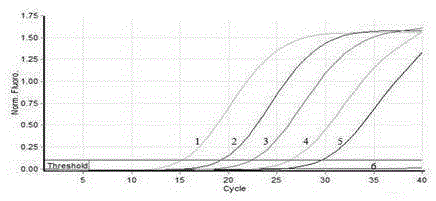
[0063] Rabbit Eimeria medus 10 6 Make an oocyst DNA stock solution at a ratio of 1:10 to 1:10 6 Doubling dilution, detect with the real-timePCR method that embodiment 2 establishes, analysis sensitivity result sees image 3 , the DNA of at least 10 oocysts can be detected.
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com