Microsatellite multiplex-PCR (Polymerase Chain Reaction) method for carrying out paternity testing on crassostrea gigas
A technology for paternity testing and oyster growth, which is applied in biochemical equipment and methods, microbiological measurement/inspection, etc., can solve the problems of easy loss and change of physical markers, low reliability, etc., and achieve the effect of improving analysis accuracy and experimental efficiency
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment
[0031] (1) Select 1-year-old long oyster broodstock with mature gonads for artificial induction, and establish 12 full-sibling single-pair mating families; collect all parent individuals (24 in total) and some offspring individuals (40 for each family) from each family. D-shaped larva);
[0032] (2) use the phenol chloroform method to extract the DNA of each parent, and use the chelating resin method to extract the DNA of each larva;
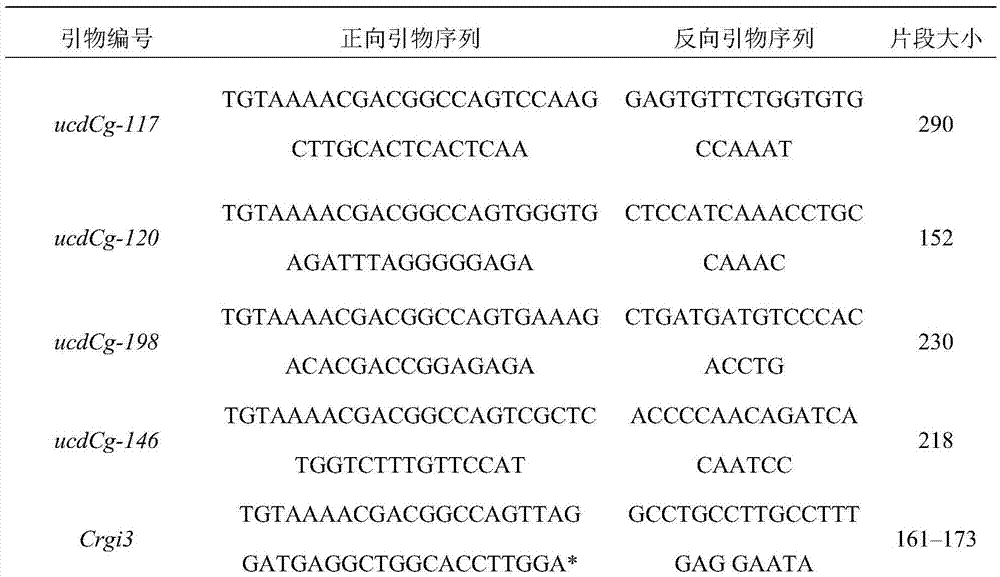
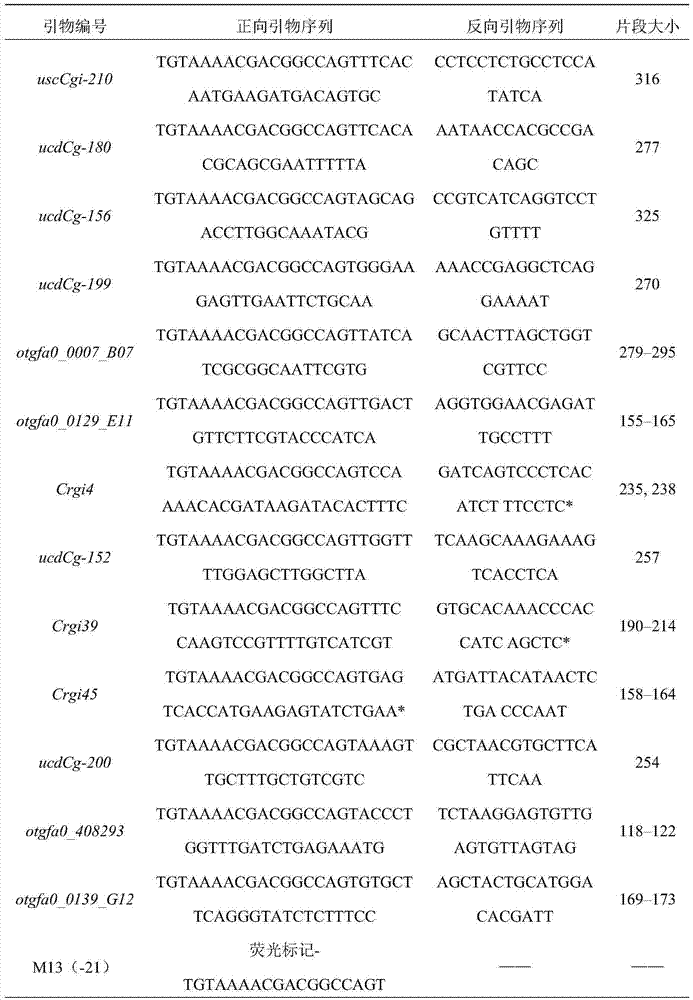
[0033] (3) Synthesize primers according to the 18 primer sequences and universal primer sequences screened above;
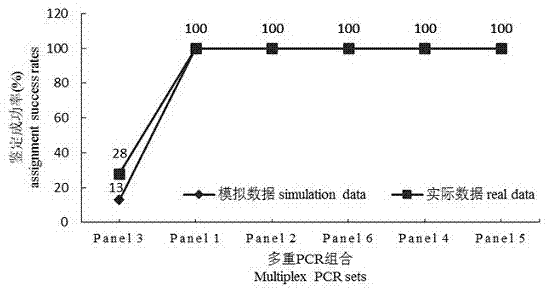
[0034] (4) Multiplex PCR amplification was carried out using the aforementioned 6 kinds of primer combinations, and the reaction system of multiplex PCR was shown in Table 1;
[0035] The reaction program of multiplex PCR was: 94°C for 3min; denaturation at 94°C for 30s, annealing at annealing temperature for 60s, extension at 72°C for 75s, 35 cycles; denaturation at 94°C for 30s, annealing at 53°C for 60s, extension at 72°C for 7...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com