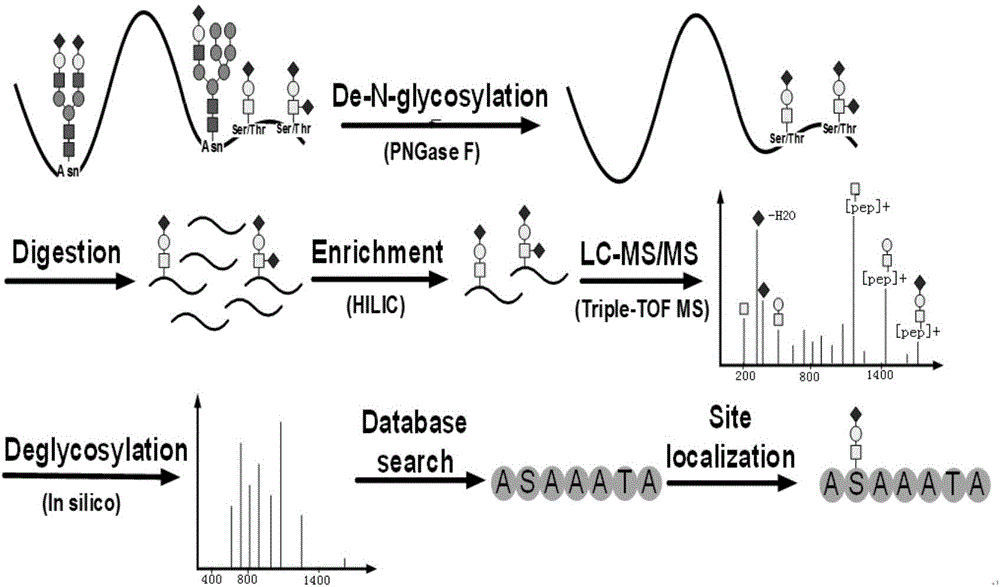
Identification method for O-glycosylation peptide fragment and complete saccharide chain thereof
An identification method and glycosylation technology, which is applied in the direction of measuring devices, instruments, scientific instruments, etc., can solve the problems of fragmentation of the peptide sequence skeleton, poor spectrum quality, neutral loss, etc., and achieve simple and fast operation and high sensitivity High, the effect of improving sensitivity
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0011] O-glycosylation analysis of embodiment 1 bovine fetuin:
[0012] 1. Add 100 μg bovine fetuin standard sample to a 0.5mL ultrafiltration tube with a molecular weight cut-off of 10kDa, add 300μL 8M urea / 100mM NH 4 HCO 3 , ultrafiltered at a centrifugal force of 14000g for 15 minutes, and with 300μL 8M urea / 100mMNH 4 HCO 3 Repeat ultrafiltration once.
[0013] 2. Add 10 μL of 1MDTT to the above ultrafiltration tube, react at 37°C for 2 hours, then add 3.8 mg of IAA, and react for 40 minutes at room temperature in the dark. Remove excess DTT or IAA by ultrafiltration for 15 min, and wash with 400 μL HO 2 O was washed twice, ultrafiltration was performed for 15 minutes each time, and the centrifugal force was 14000g.
[0014] 3. After the ultrafiltration is completed, add PNGaseF glycosidase 2 μL to the protein sample in the above ultrafiltration tube, and react overnight at 37°C. After the reaction was completed, ultrafilter again, and add 400 μL HO 2 O was washed tw...
Embodiment 2
[0022] O-glycosylation analysis of embodiment 2 human serum albumin
[0023] 1. Add 60 μL of human serum sample to a 0.5 mL ultrafiltration tube with a molecular weight cut-off of 10 kDa, add 300 μL of 8M urea / 100mM NH 4 HCO 3 , ultrafiltered at a centrifugal force of 14000g for 15 minutes, and with 300μL 8M urea / 100mMNH 4 HCO 3 Repeat ultrafiltration once;
[0024] 2. Add 10 μL of 1MDTT to the above ultrafiltration tube, react at 37°C for 2 hours, then add 3.8 mg of IAA, and react for 40 minutes at room temperature in the dark. Remove excess DTT or IAA by ultrafiltration for 15 min, and wash with 400 μL HO 2 O was washed twice, ultrafiltration was performed for 15 minutes each time, and the centrifugal force was 14000g.
[0025] 3. After the ultrafiltration is completed, add PNGaseF glycosidase 2 μL to the protein sample in the above ultrafiltration tube, and react overnight at 37°C. After the reaction was completed, ultrafilter again, and add 400 μL HO 2 O was washed ...
PUM
| Property | Measurement | Unit |
|---|---|---|
| particle diameter | aaaaa | aaaaa |
Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com