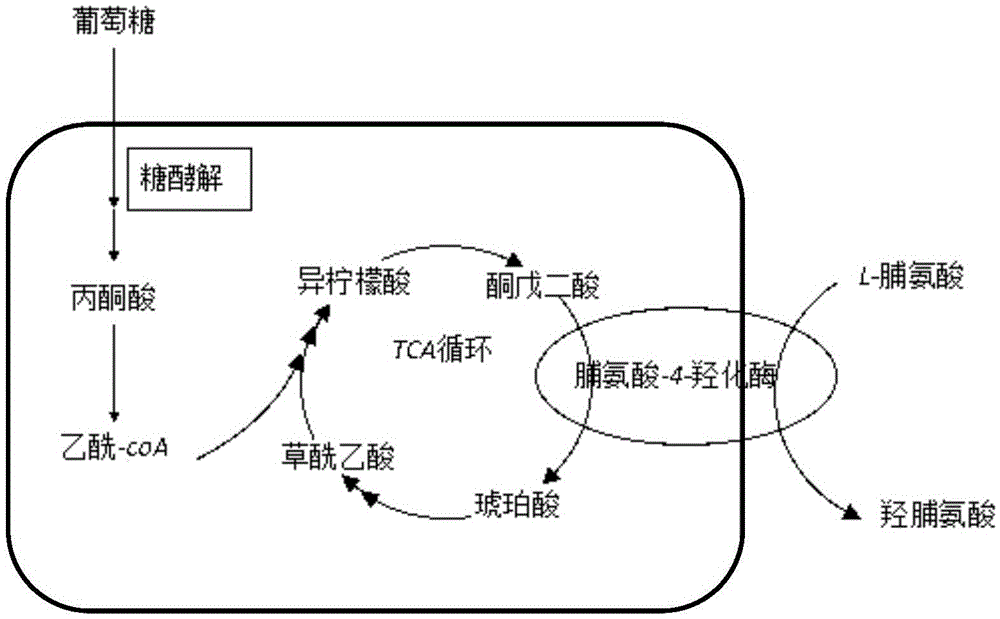
Biosynthesis method for increasing yield of trans-4-hydroxyproline by knocking out other metabolic pathway
A technology of hydroxyproline and metabolic pathways, applied in the field of microbial genetic engineering, can solve the problems of waste of raw materials, high cost, and many synthesis steps
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
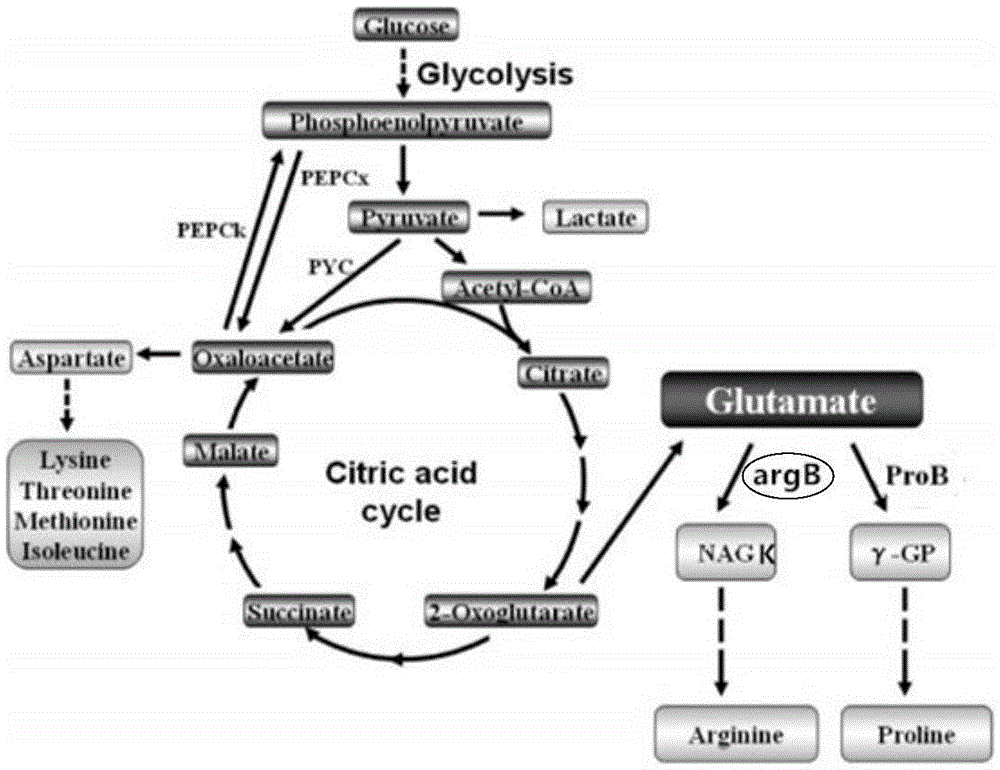
[0026] Example 1: argB gene knockout
[0027] The gene sequence of argB obtained from NCBI (GenBank: AM946981.2): argB gene (SEQ ID NO: 1)
[0028] Using short homology arms, design primer pairs: primer 5'-3': P1 (SEQIDNO: 2), P2 (SEQIDNO: 3)
[0029] Using pKD4 as a template, a target fragment containing the upstream and downstream homology arms of argB and the resistance gene (Kan) was obtained. The E. coli BL21(DE3) △putA containing plasmid pKD46 was obtained by chemical transformation, and then it was prepared as electro-competent, and the target fragment was introduced into the electro-competent by electroporator. The effect of the three proteins expressed in plasmid pKD46 Next, use the Red recombination system to complete gene recombination. After electroporation, resuscitate at 30°C for 3 hours, and spread a plate containing kanamycin. After that, the plasmid pCP20 was transferred to eliminate the resistance gene. The strains were spotted on non-resistant, Amp resistant, an...
Embodiment 2
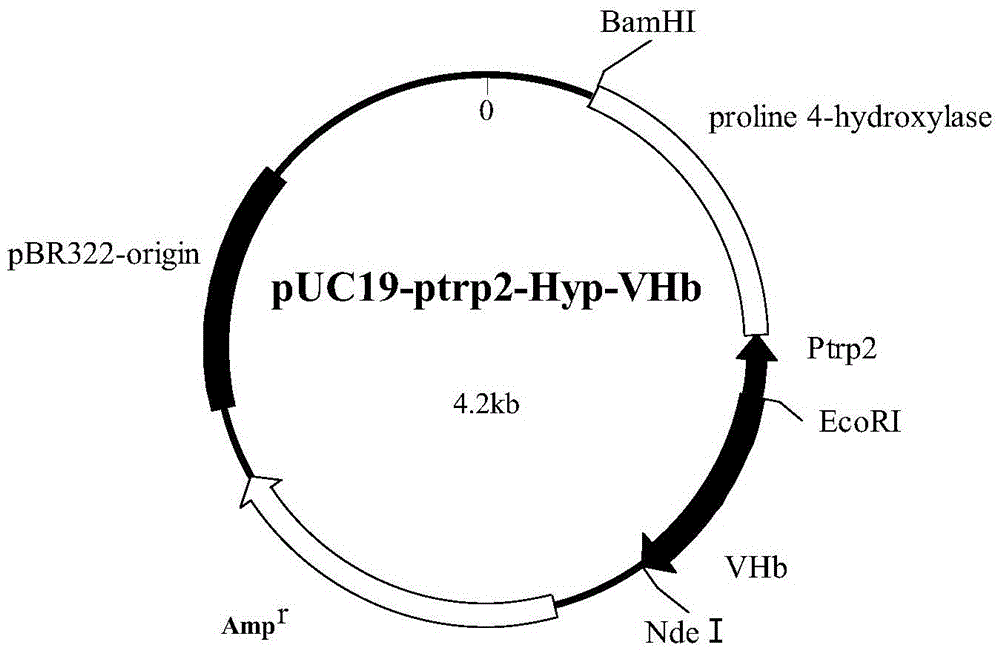
[0030] Example 2: Obtaining the recombinant plasmid (pUC19-ptrp2-Hyp-VHb)
[0031] Activate the strain containing the target plasmid stored in the laboratory and incubate at 37°C and 220 rpm for 12-16 hours. Take the cultured bacterial liquid to extract the plasmid, and all operations are carried out in strict accordance with the instructions.
Embodiment 3
[0032] Example 3: Construction of recombinant strains
[0033] Take out the E. coli BL21(DE3) △putA / △argB competent, place it on ice and thaw, add 37μL of sterile water, 10μL KCM, 3μL plasmid. After mixing, place on ice for 30min, heat shock at 42°C for 90s, then place on ice for 5min, add 600μL of fresh LB medium, culture with shaking at 37°C for 1h, take 100μL and spread on ampicillin resistant plate. After culturing at 37°C for 8-16 hours, when a single colony grows, select a single colony for verification.
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com