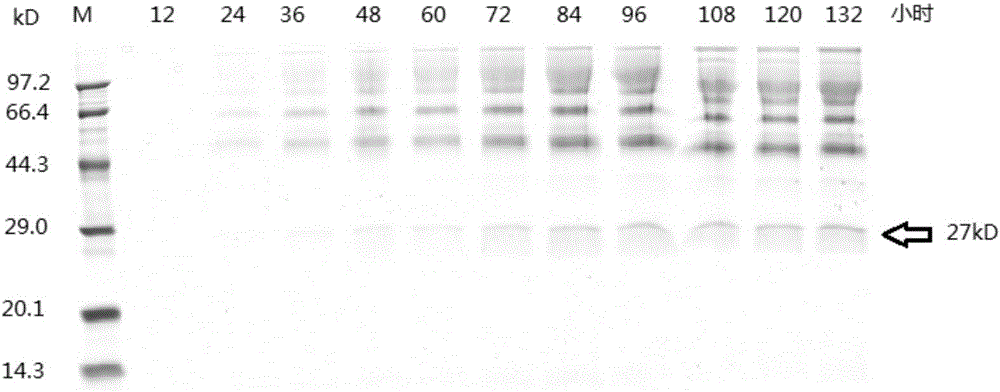
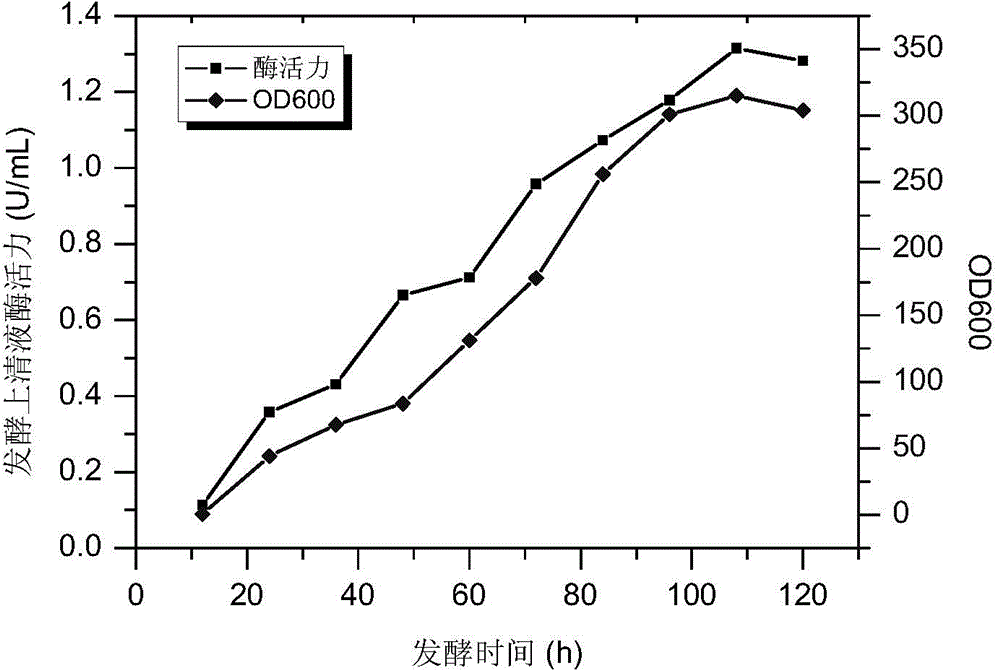
Constitutive expression and purification of ampullaria gigas endogenous cellulase EG27I in pichia yeast
A technology of cellulase and snails, applied in the direction of enzymes, enzymes, recombinant DNA technology, etc., can solve the problems of difficulty in recombinant expression of EG27I
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1p
[0019] The construction of embodiment 1 pGAPH vector
[0020] Synthesize a piece of dsDNA as follows:
[0021] 5'-ACTCAATTGAACAACTATTTCGCCACCATGCAATTCTTCGCTGTTGCTTTGTTCGCTACTAGTGCTTTGGCTGAATTCGGTACCAAT-3', the dsDNA was digested with MfeI and KpnI restriction enzymes, and the digested product purified by the product purification kit was ligated with the pGAPZαA vector (InvitrogenCo., Ltd.) digested with the same enzyme, ligated The reaction system is as follows:
[0022]
[0023] The above mixture was incubated at 22°C for 4 hours. The resulting ligation product was transformed into E.coliDH5α competent cells, coated with bleomycin resistance plates, positive clones were screened, and plasmid DNA was prepared using a plasmid extraction kit. The resulting plasmid DNA was digested with NcoI and SpeI to preliminarily identify the plasmid pGAPH.
Embodiment 2
[0024] Construction of embodiment 2 expression plasmid pGAPH-EG27I
[0025] First design the following primer pairs:
[0026] Upstream: 5'-GGC ACTAG TGCTTTGGCTGCACAGTTGTGTCAG-3'
[0027] Downstream: 5'-CGC TCTAGA TTAGCCCGAATTGTGGCATTCAC-3'
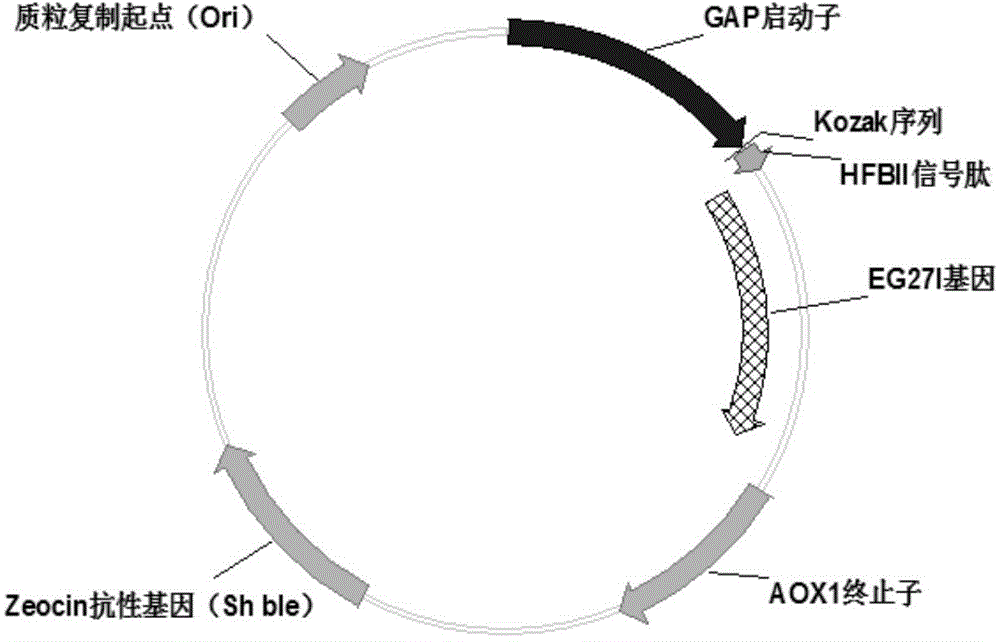
[0028] Using pPIC9K-eg27I [RuiGuoetal.ActaBiochimBiophysSin, 2008:419-425] as a template, using the above primer pair, PCR amplified the gene of EG27I, after the product was purified and recovered the target fragment, it was digested with SpeI and XbaI and recovered. The pGAPH plasmid is connected, and the connection conditions are the same as in Example 1 to obtain the plasmid pGAPH-EG27I, and its structural representation is as follows figure 1 As shown, its core region contains GAP promoter, Kozak sequence, HFBII signal peptide and target gene EG27I. The ligation product was transformed into E.coliDH5α competent cells, and the positive clones were screened on a bleomycin-resistant plate. The plasmid DNA was prepared by alkaline ...
Embodiment 3
[0029] Example 3 Transformation of Pichia pastoris SMD1163 with expression plasmid
[0030]The constructed recombinant expression plasmid pGAPH-EG27I was digested with BlnI and linearized, recovered by phenol / chloroform extraction and dissolved in sterile water. According to the method introduced in the Invitrogen manual [InvitrogencorpSD, CA. Amanual of methods expression of recombinant proteins in piciapastoris, 1998] to prepare SMD1163 electrotransformed cells, mix the above 10 μg linearized plasmid with 80 μl electrotransformed cells, and use Bio-Rad electroporator for electroporation transformation. The electroporation conditions of Saccharomyces cerevisiae were set. Transfer the electroporation product to a 10ml sterile centrifuge tube, immediately add 1ml of 1mol / L sorbitol pre-cooled at 0°C, let stand at 30°C for two hours, add 2ml of YPD (10g / LYeastExtract, 20g / LTryptone, 20g / LD-glucose) Incubate overnight at 30°C. The next day, the supernatant was removed by quick ...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com