Novel mutant pathogenic gene PLEKHM1 of osteopetrosis, and encoded protein and application thereof
A technology of PLEKHM1 and osteopetrosis, applied in the field of medical molecular biology, can solve incomplete problems and achieve the effect of enriching the resource base and effective molecular diagnosis basis
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0052] Example 1 Obtaining and Identification of Mutant Gene PLEKHM1
[0053] 1. Sample Collection
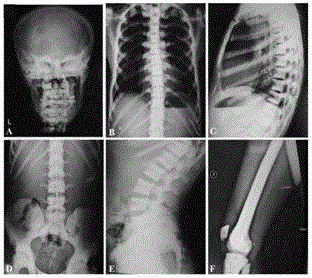
[0054] The present inventor firstly carried out interrogation, general laboratory examination and bone imaging examination to the patient with osteosclerosis, and found that the patient had symptoms such as easy fracture, tooth loss, anemia, hepatosplenomegaly, and the bone X-ray photograph showed skull, Vertebrae, long bones, etc. have extensive bone density increase. Vertebral bone biopsy pathological smears show bone marrow cavity narrowing and hematopoietic tissue decrease, which are generally consistent with the pathological features of osteosclerosis (see appendix figure 1 ,2,3).
[0055] 2. DNA extraction
[0056] The peripheral blood of the proband and his family members was collected, and the whole genome DNA was extracted from the peripheral blood using a kit (TIANGEN BIOTECH, Beijing, China, DP304-03). The DNA concentration was measured by ultraviolet spectrophotom...
Embodiment 2
[0066] Construction of embodiment 2 expression vector
[0067] The wild-type PLEKHM1 protein expression vector (FLAG-PLEKHM1-WT) was provided by Professor Tamotsu Yoshimori of Osaka University in Japan. The inventor applied the overlap extension PCR method to construct the mutant PLEKHM1 protein expression vector (FLAG-PLEKHM1-MU). The vector plasmid backbone used For FLAG-HA-pcDNA3.1, the details are as Figure 5 shown. First, using the FLAG-PLEKHM1-WT plasmid as a template, the full-length sequence of PLEKHM1 was amplified by overlapping extension PCR technology, and the mutation was introduced into the target fragment, and then the target fragment with the mutation was combined with the vector, and the target gene was introduced into the recipient cells. Escherichia coli DH5α was used for screening and cultivation, and the target gene was obtained from the flora. It is a common technique in this field.
[0068] After identification, the mutant PLEKHM1 protein expression ...
Embodiment 3
[0069] Embodiment 3 Mutant protein expression and identification of function
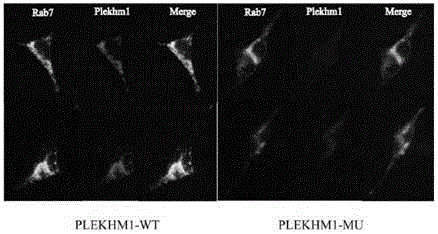
[0070] PLEKHM1 protein can be combined with Rab7 protein, the wild-type and mutant proteins were expressed by HEK293 cells, and then immunofluorescence was used to detect related proteins, the results Figure 7 shown. Green shows Rab7 protein, red shows PLEKHM1 protein, when the two are fused, they show yellow. The wild-type PLEKHM1 protein is on the left, and the mutant PLEKHM1 protein is on the right. By comparison, it can be found that the expression of PLEKHM1 protein is reduced after mutation, and the combination with Rab7 protein is reduced.
[0071] Co-immunoprecipitation technique was used to detect the combination of wild-type and mutant PLEKHM1 protein and Rab7 protein, such as Figure 8 shown. The Rab7 protein and the wild-type or mutant PLEKHM1 protein were transfected into HEK293 cells, and the cells were lysed after 24 hours. First, immunoprecipitation was performed with FLAG-speci...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com