Clone and application of key gene PeIRX10 for phyllostachys edulis xylan synthesis
A key gene, xylan technology, applied in the field of cloning and functional exploration of the key gene PeIRX10 of moso bamboo glycosyltransferase
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
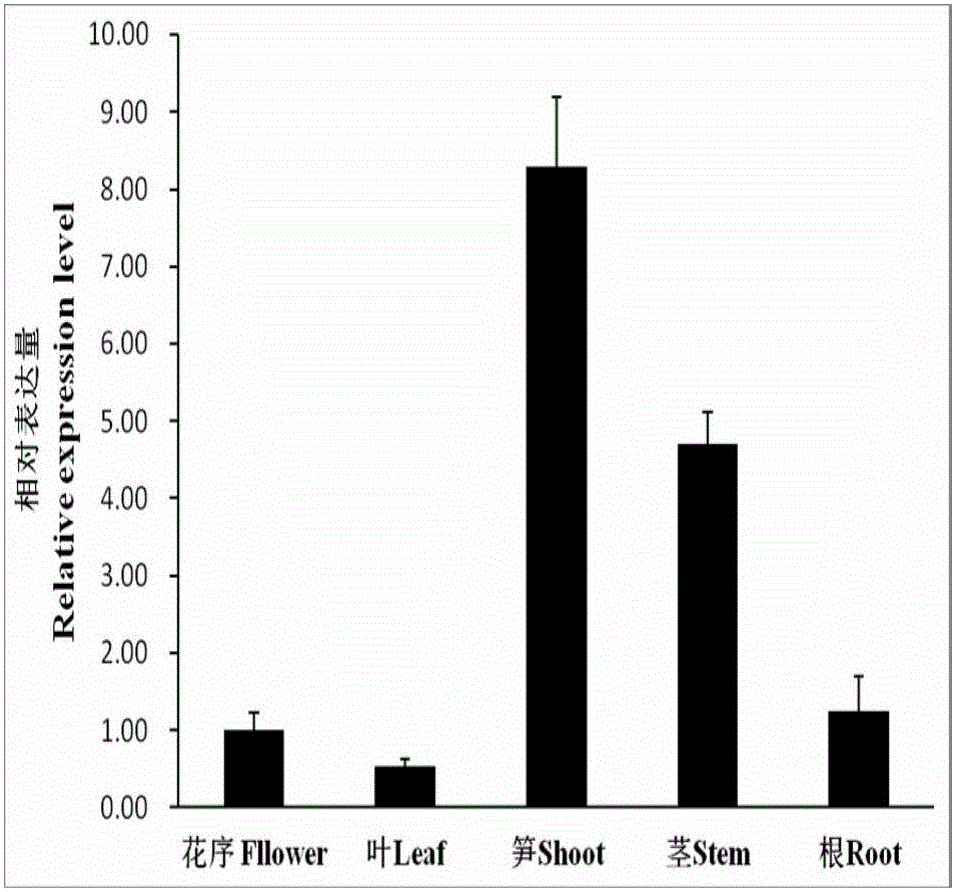
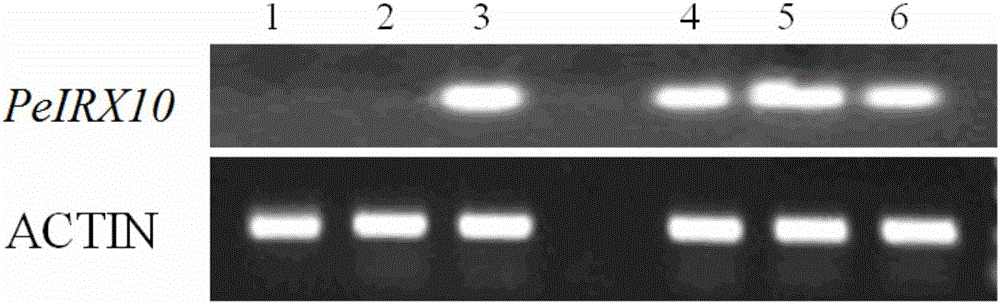
[0023] Example 1 Phyllostachys pubescens PeIRX10 gene expression
[0024] (1) Experimental method
[0025] 1. Moso bamboo material
[0026] Moso bamboo materials of different tissues were obtained from the roots, stems, leaves, inflorescences and young shoots of 1-year-old seedlings, which were quick-frozen in liquid nitrogen and stored at -80°C for the extraction of total RNA.
[0027] (1) RNA extraction and cDNA synthesis
[0028] Perform BLAST retrieval and comparison analysis on the sequenced gene sequence database and its corresponding protein sequence database in Phyllostachys pubescens, obtain the nucleotide sequence of PeIRX10 gene (PH01004923G0080; SEQ ID No: 1), and design primers as follows:
[0029] PeIRX10-F1:5'-CCTGAACCACATGTTTGCCG-3' (SEQ ID No: 2)
[0030] PeIRX10-R1:5'-AATCGCACTGCGCATCATTC-3' (SEQ ID No: 3)
[0031] The total RNA of each sample of Phyllostachys pubescens was extracted with RNA extraction kit (OMEGA). Reverse transcription was performed us...
Embodiment 2
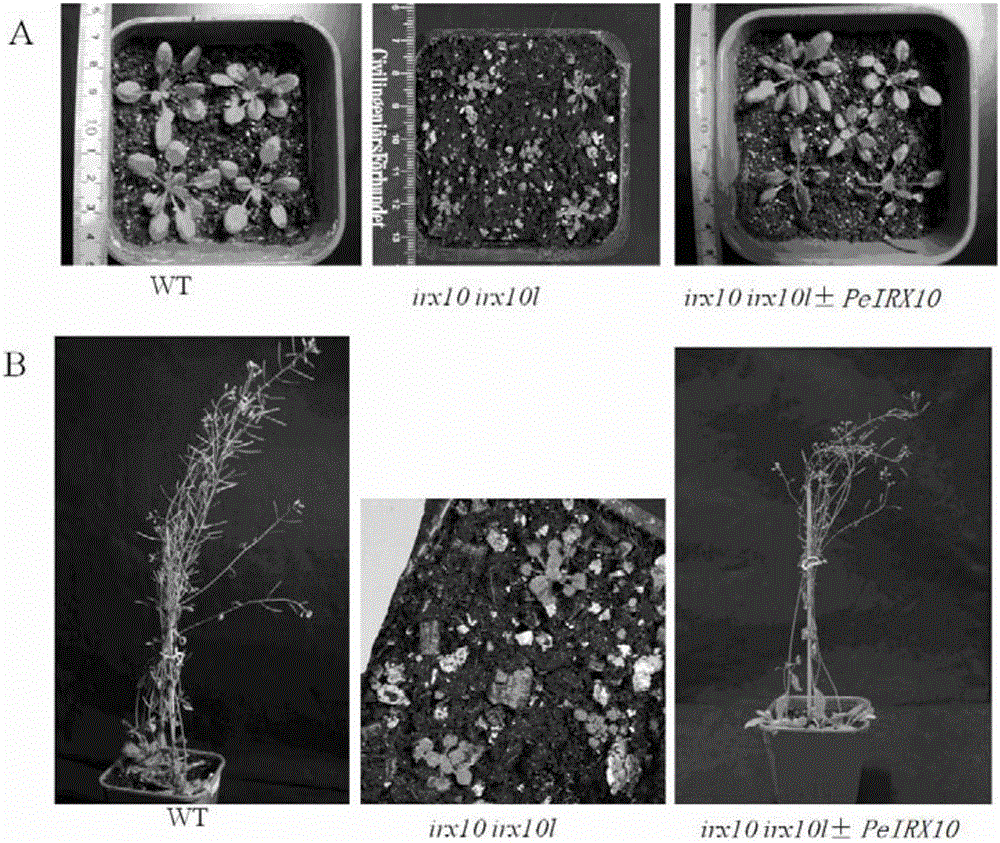
[0039] Example 2 Overexpression of Phyllostachys pubescens PeIRX10 gene in Arabidopsis thaliana
[0040] (1) Experimental method
[0041] 1. Construction of expression vector
[0042] Based on the Gateway system for PCR amplification of the full-length cDNA sequence of Phyllostachys pubescens PeIRX10, the amplification primers are:
[0043] PeIRX10-F:
[0044] 5′-
[0045] GGGGACAAGTTTGTACAAAAAAGCAGGCTTCATGAGGAGGTGGGTCTTGGCC-3' (SEQ ID No: 6);
[0046] PeIRX10-R:
[0047] 5′-
[0048] GGGGACCACTTTGTACAAGAAAGCTGGGTCCCAAGGCTTCAGGTCGCCCACCG-3' (SEQ ID No: 7).
[0049] The PCR reaction system is 20 μL: 10 μL of 2×PrimeSTAR Max Premix, upstream and downstream primers (10 μmol L -1 ) each 0.5 μL, template 2 μL, dd H 2 O 7 μL. Reaction program: pre-denaturation at 94°C for 5 minutes; 35 cycles at 94°C for 30s, 1min at 55°C for 30s, and 30s at 72°C; extension at 72°C for 10min. The amplified product was ligated with pDONR207, transformed into DH5α, and positive clones were obt...
Embodiment 3
[0061] Example 3 Cell wall xylan immunolocalization
[0062] (1) Experimental method
[0063] Take the stems at the base of wild-type, double-horn plants and complementary Arabidopsis plants that have grown for eight weeks, cut them into 1mm-thick slices, wash the slices with 0.1M phosphate buffer (pH 7.2) for 5-10min; use fresh Soak the slices in 3% skimmed milk for 1 h and blow the skim milk continuously; remove the skim milk and wash the slices with PBS buffer for 5 min; incubate the slices with rat anti-xylan antibody LM10 (Plantprobes) for 2 h, then wash the slices with PBS buffer to Wash unbound primary antibody; incubate with 50-fold diluted FITC-goat anti-rat antibody (Zomanbio, Cat.Z1319) for 2 h, then wash the section with PBS buffer 10 times to wash unbound secondary antibody; finally The sections were fixed on glass slides, observed and photographed under a laser confocal electron microscope (Zeiss LSM710, 495nm), and the results are shown in the attached Figure...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com