Schistosoma japonicum katsurada mitochondrial molecular marker and application thereof
A technology of schistosomiasis, mitochondrial gene, applied in the field of biotechnology and genetics
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0072] Example 1 Sample Collection, Genomic DNA Extraction and Amplification of Target Fragments
[0073] (1) Collect samples of Schistosoma japonicum from 12 endemic areas including Guichi in Anhui, Tongling in Anhui, Yueyang in Hunan, Changde in Hunan, Shashi in Hubei, Duchang in Jiangxi, Xichang in Sichuan, Eryuan in Yunnan, Taiwan, Japan, the Philippines, and Indonesia, Genomic DNA of a single individual was extracted (extraction reagent: DNeasy Blood & Tissue Kit produced by QIAGEN (Germany)). The specific method is as follows:
[0074] Put a single adult Schistosoma japonicum into a 1.5ml EP tube, add 500μl of 1.0mM EDTA and 10mM Tris buffer. After oscillating, suck up the liquid and repeat three times. Add 180 μl of ATL buffer provided in the kit; add 20 μl proteinase K, shake and mix. Incubate in a water bath at 56 °C for 4 h, shaking several times in between. Shake for 15 seconds after taking it out of the water bath, add 200 μl buffer AL, and shake to mix. Then ...
Embodiment 2
[0085] Example 2 Sequence Alignment
[0086]Using MEGA 5.1 software, the gene sequence obtained by sequencing in Example 1 was used for multiple sequence alignment with the reference sequence (database) of known geographical origin in the standard database.
[0087] One of the sequence alignment results (ND1 gene) can be seen figure 1 , it can be seen that there are quite a lot of base variations among individuals in different endemic areas. The other two genes also had base variations among different individuals (results not shown).
Embodiment 3
[0088] Construction of embodiment 3 phylogenetic tree
[0089] Through model analysis software (jmodeltest) evaluation and manual sampling inspection, the most suitable nucleotide evolution model was selected as the HKY model, and the evolutionary tree was constructed based on the selected model by the maximum likelihood method (Maximum Likelihood, ML). According to the topological structure of the phylogenetic tree (especially each branch point and Bootstrap value), the source region of the sample was judged.
[0090] 3.1 The phylogenetic tree construction of a single gene
[0091] 3.1.1 Phylogenetic tree constructed by ND1
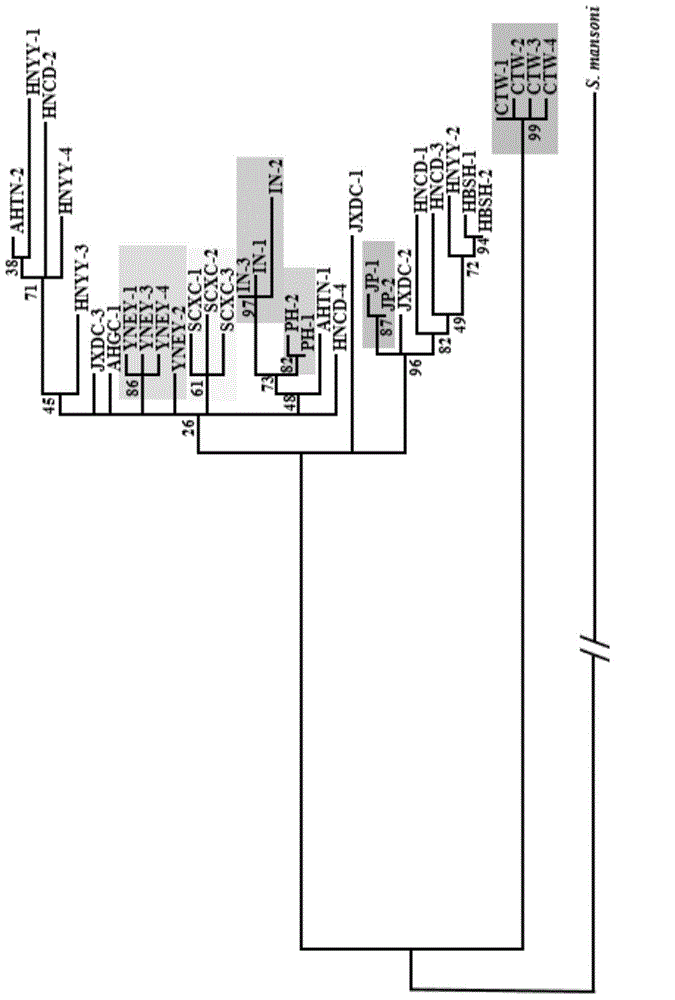
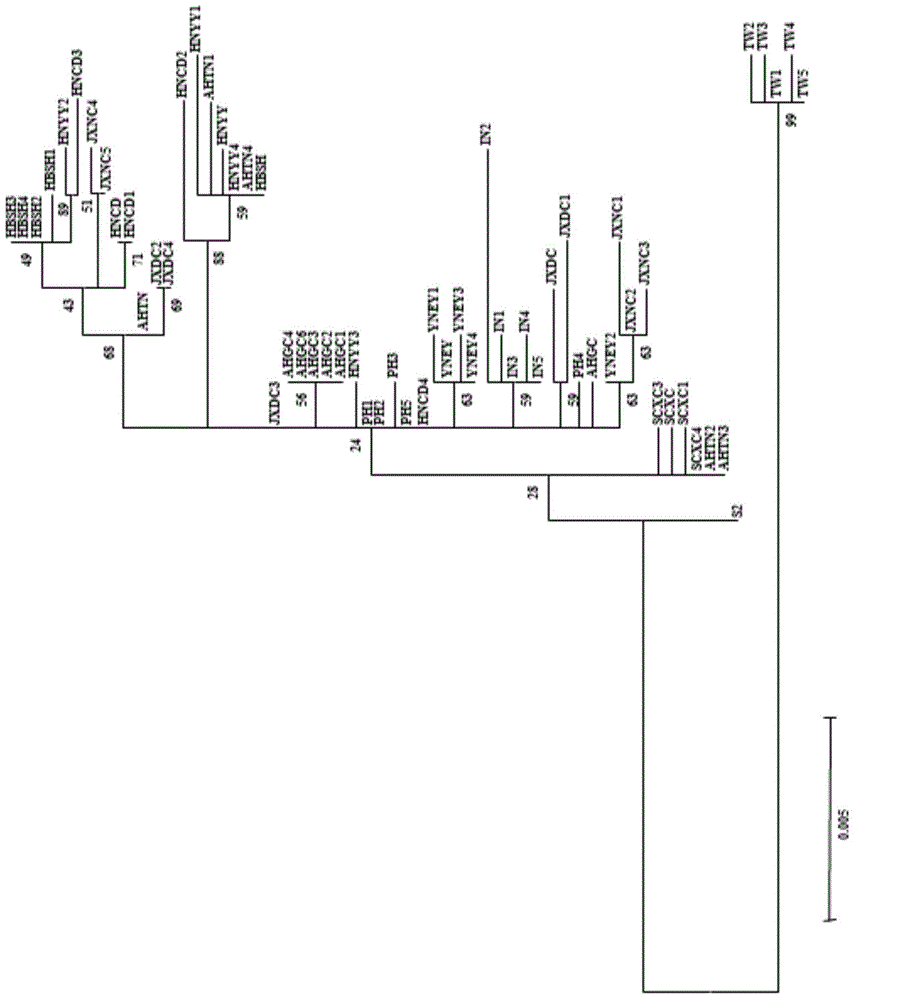
[0092] ML phylogenetic tree constructed with ND1 gene, see the results image 3 , it can be seen that the ND1 gene can distinguish Taiwan (TW), Indonesia (IN) and Yunnan (YNEY) very well;
[0093] 3.1.2 Phylogenetic tree constructed by ND4
[0094] ML phylogenetic tree constructed with ND4 gene, see the results Figure 4 , it can be seen that the ND...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com