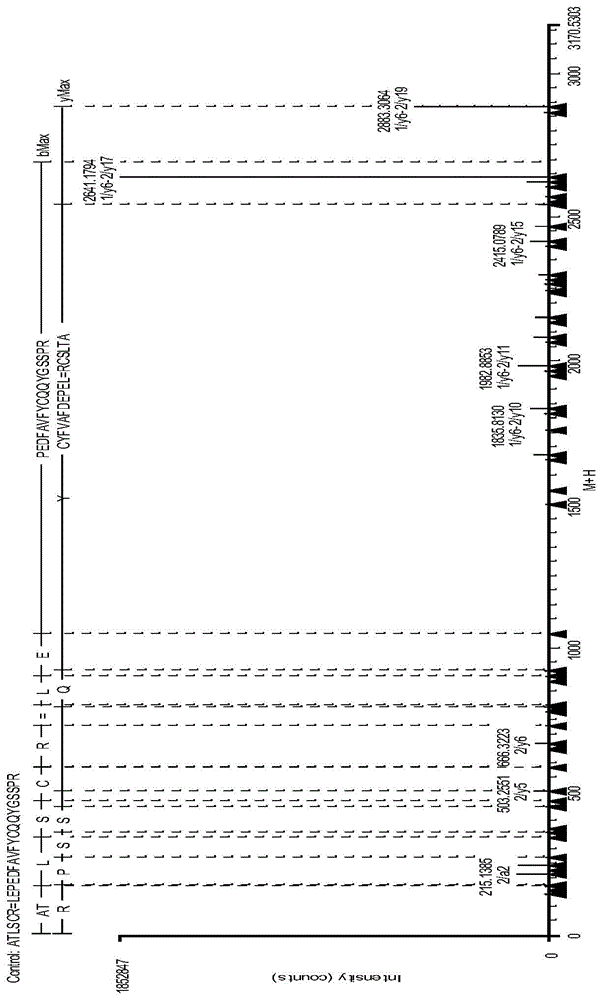
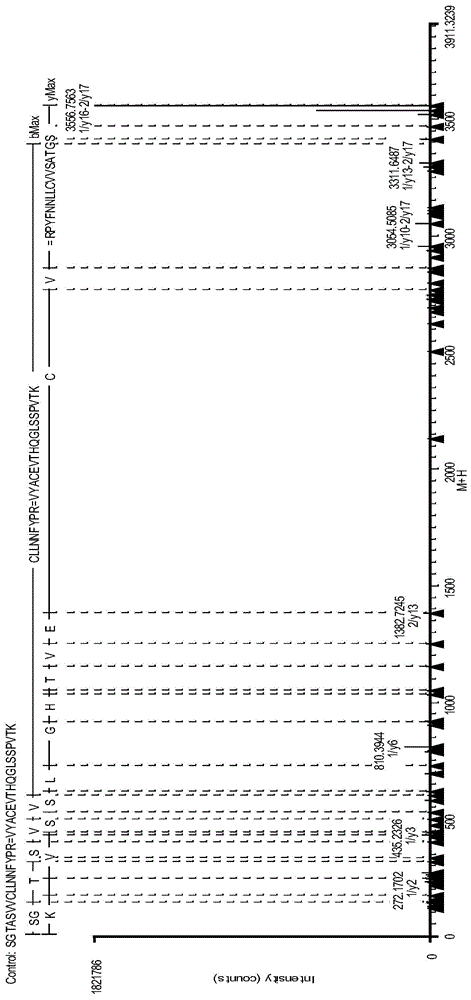
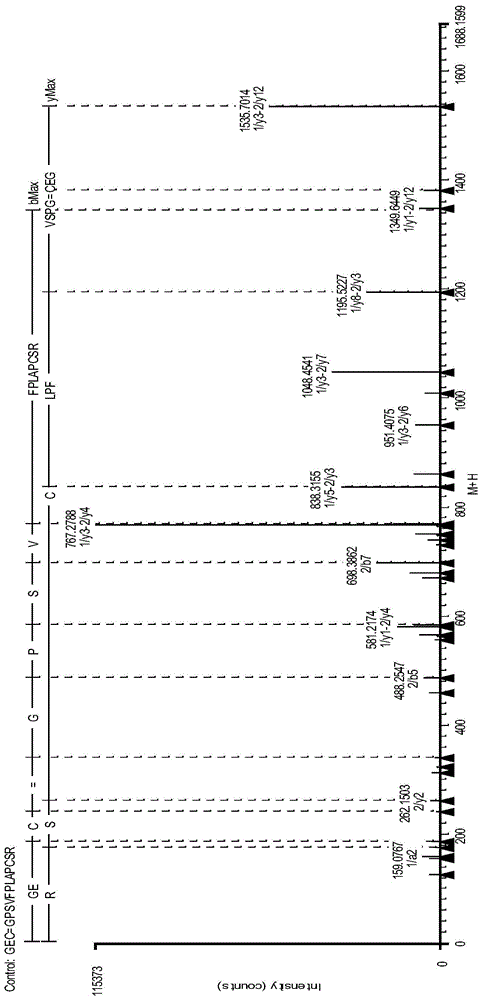
IgG2 type monoclonal antibody disulfide bond pairing analysis method
An analysis method, disulfide bond technology, applied in the field of monoclonal antibody disulfide bond pairing analysis, can solve inconvenience and other problems, and achieve the effect of enriching fragment ions
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0050] Example 1: Antibody Protein Disulfide Bond Pairing Analysis (Trypsin Digestion)
[0051] Antibody protein disulfide bond pairing analysis steps:
[0052] (1) Protein denaturation treatment
[0053] Take 0.5 mg protein sample and add it to 380 μl denaturation buffer (8M guanidine hydrochloride+5mM EDTA+0.5MTris, pH8.3), mix well, and incubate at 37°C for 60 minutes. Ultrafiltration centrifugation or desalting column desalting to 50mM NH 4 HCO 3 (pH8.0) buffer.
[0054] (2) Enzymolysis
[0055] Take 100 μg of the protein in (1), add trypsin to digest the antibody protein at a ratio of 1:10 (wt / wt, enzyme / protein), and incubate at 37°C for 4 hours.
[0056] (3) Reduction and termination of enzymatic hydrolysis
[0057] After the enzymatic hydrolysis of the sample in (2) is completed, take out half of the sample volume and add formic acid FA to a final volume concentration of 0.1%; add 1 μL of 0.1M DTT (dithiothreitol) to the other half of the sample volume. Formic a...
Embodiment 2
[0075] Example 2: Analysis of antibody protein disulfide bond pairing (trypsin 1:25 digestion)
[0076] Antibody protein disulfide bond pairing analysis steps:
[0077] (1) Protein denaturation treatment
[0078] Take 0.5 mg protein sample and add it to 380 μl denaturation buffer (8M guanidine hydrochloride+5mM EDTA+0.5MTris, pH8.3), mix well, and incubate at 37°C for 60 minutes. Ultrafiltration centrifugation or desalting column desalting to 50mM NH 4 HCO 3 (pH8.0) buffer.
[0079] (2) Enzymolysis
[0080] Take 100 μg of the protein in (1), add trypsin to digest the antibody protein at 1:25 (wt / wt), and incubate at 37°C for 4 hours.
[0081] (3) Reduction and termination of enzymatic hydrolysis
[0082] After the enzymatic hydrolysis of the sample in (2) is completed, take out half of the sample volume and add formic acid FA to a final concentration of 0.1%; add 1 μL of 0.1M DTT to the other half of the sample volume, and add formic acid FA to a final concentration of 0...
Embodiment 3
[0087] Example 3: Analysis of antibody protein disulfide bond pairing (trypsin combined with chymotrypsin digestion)
[0088] Antibody protein disulfide bond pairing analysis steps:
[0089] (1) Protein denaturation treatment
[0090] Take 0.5 mg protein sample and add it to 380 μl denaturation buffer (8M guanidine hydrochloride+5mM EDTA+0.5MTris, pH8.3), mix well, and incubate at 37°C for 60 minutes. Ultrafiltration centrifugation or desalting column desalting to 50mM NH 4 HCO 3 (pH8.0) buffer.
[0091] (2) Enzymolysis
[0092] Take 100 μg of the protein in (2), add trypsin at 1:25 (wt / wt), add chymotrypsin combined with enzymatic antibody protein at 1:50 (wt / wt), and incubate at 37°C for 4 hours.
[0093] (3) Reduction and termination of enzymatic hydrolysis
[0094] After the enzymatic hydrolysis of the sample in (2) is completed, take out half of the sample volume and add formic acid FA to a final concentration of 0.1%; add 1 μL of 0.1M DTT to the other half of the s...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com