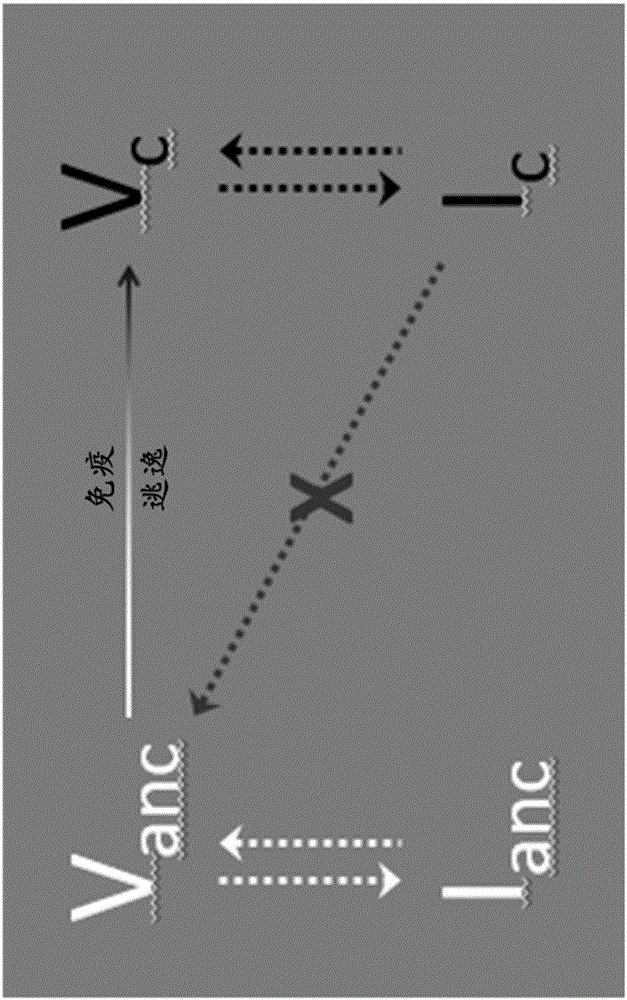
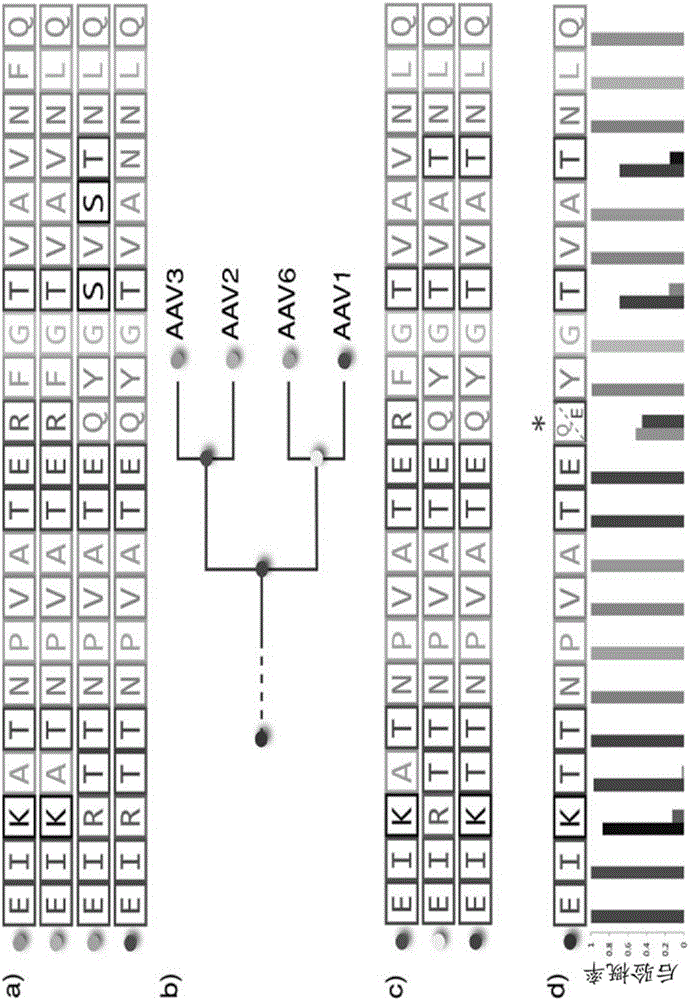
Methods of predicting ancestral virus sequences and uses thereof
A sequence, ancestral technology, applied in the direction of viruses, viral peptides, viruses/phages, etc., can solve problems such as reducing efficiency and increasing safety risks by gene transfer
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
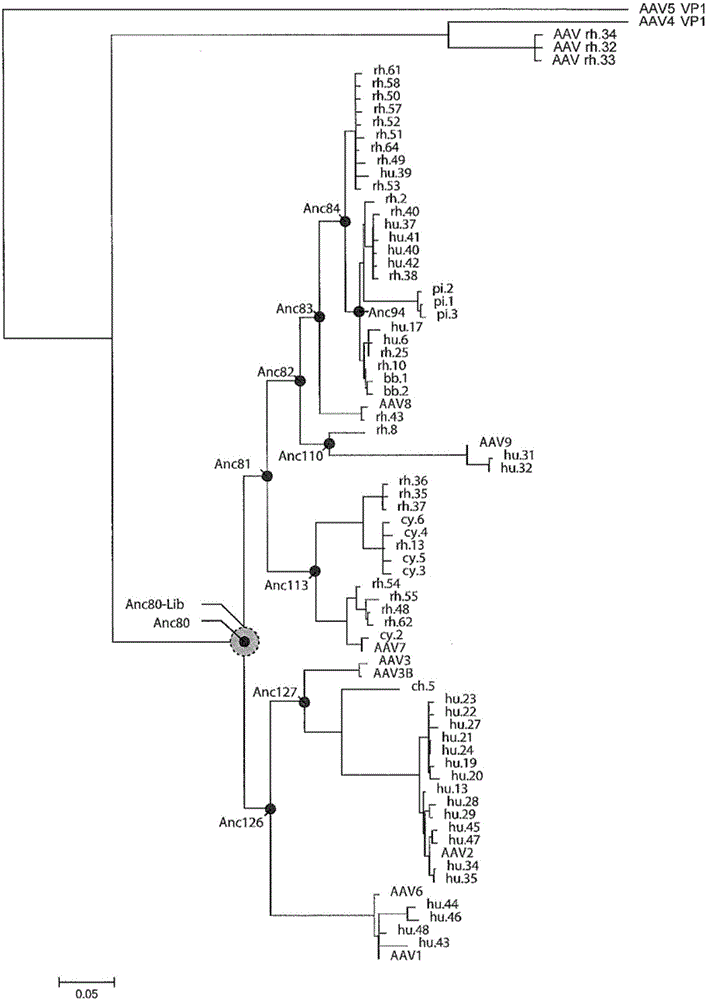
[0093] Example 1: In silico prediction of ancestral sequences
[0094] A set of 75 different amino acid sequences for AAV capsids were obtained from a number of public databases, including GenBank, and the sequences were aligned using the PRANK-MSA algorithm, version 121002, with option "-F".
[0095] ProtTest3 (see e.g. Darriba et al., 2011, Bioinformatics, 27(8):1164-5; available on the World Wide Web at darwin.uvigo.es / software / prottest3) was used under different conditions (e.g. in ProTest3 Those conditions included, i.e. "+I", "+F", "+G" and their combinations) evaluate different models of peptide evolution (e.g. those included in ProTest3, i.e. JTT, LG, WAG, VT, CpRev, RtRev, Dayhoff, DCMut, FLU, Blosum62, VT, HIVb, MtArt, MtMam). The JTT model was selected based on the Aikake Information Criterion (AIC; Hirotugu, 1974, IEEE Transactions on Automatic Control, 19:716-23) score as implemented in ProTest3 (Jones et al., 1992, Comp. Appl. Biosci., 8: 275-82) and +G and +F ...
Embodiment 2
[0107] Example 2: Expression of Ancestral AAV VP1 Sequences
[0108] Experiments were performed to determine whether the predicted ancestral AAV capsid sequences could be used to make viral vectors.
[0109] A number of predicted ancestral AAV capsid sequences were cloned. Transfer of the ancestral capsid library to the rep-cap expression plasmid enables viral particle formation in a transient transfection. To maintain proper expression levels and splicing of VP1, VP2 and VP3, by cleaving HindIII located 5' of cap in the rep coding sequence and SpeI engineered between the cap stop codon and polyadenylation signal, The library cap genes were cloned. Therefore, to clone the ancestral capsid into the more conventional "REP / CAP" construct, the passage plasmid was digested with HindIII and SpeI, gel purified, and ligated into a similarly digested rep / cap plasmid.
[0110] The expressed polypeptides were resolved on 10% SDS gels. Such as Figure 6 As shown in , capsid polypepti...
Embodiment 3
[0111] Example 3: Virus titration
[0112] AAV was produced in HEK293 cells by transient co-transfection of plasmids encoding all elements required for virion assembly. Briefly, HEK293 cells were grown to 90% confluency and transfected with (a) a viral genome plasmid encoding a luciferase transgene (expressed through a CMV promoter) flanked by AAV2 ITRs, (b) encoding AAV packaging plasmids for AAV2rep and synthetic capsid proteins disclosed herein, (c) AAV2-AAP expression capsid, and (d) adenoviral helper genes required for AAV packaging and assembly. Cells were cultured at 37°C for 2 days, and cells and media were harvested.
[0113] The cell culture medium suspension was lysed by 3 consecutive freeze-thaw cycles. Afterwards, the lysate was clarified by centrifugation and treated with an enzyme (herein Benzonase TM ) to digest any DNA present outside the virus particle. Dilute the AAV prep to fall within the linear measurement range of the control DNA template, in this ca...
PUM
| Property | Measurement | Unit |
|---|---|---|
| diameter | aaaaa | aaaaa |
Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com