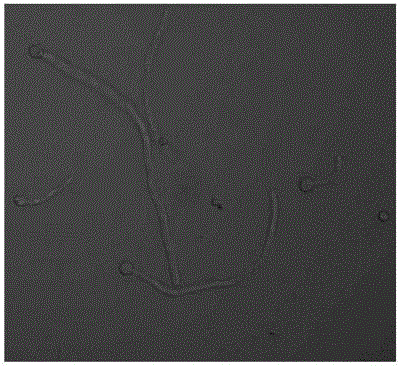
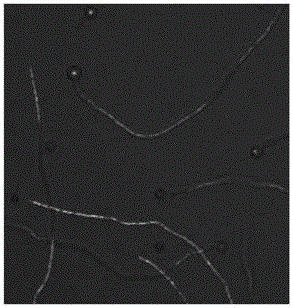
Method for expressing protein in aspergillus niger resting spores through external-source single strand RNA
A technology of Aspergillus niger spores and dormant spores, applied in the field of protein expression, can solve problems such as inability to apply, and achieve the effects of eliminating cathode effect, avoiding killing cells, and improving cell survival rate.
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0069] Expression of green fluorescent protein (GFP) in Aspergillus niger cells:
[0070] 1. Plasmid construction
[0071] Using the GFP gene coding sequence, the GFP gene is shown in SEQ ID NO.5,
[0072] The protein sequence of the above-mentioned GFP gene is shown in SEQ ID NO.6,
[0073] PCR primers for amplifying GFP gene:
[0074] F: The wavy line is the Aat II restriction site
[0075] R: The wavy line is the Sal I restriction site
[0076] The underlined GCTAGC sequence in primer F is used to promote the transcription of the DNA into RNA, and the translation initiation factor binds to the RNA. This sequence is next to the initiation codon ATG of the expressed target gene. After the above-mentioned pair of primers are used for PCR, the upstream and downstream of the GFP gene can be carried with GCTAGC, and both upstream and downstream can carry restriction sites.
[0077]
[0078] After the PCR product was detected by agarose gel electrophoresis, the PCR product was recovered a...
Embodiment 2
[0108] Expression of red fluorescent protein (RFP) in Aspergillus niger cells:
[0109] 1. Plasmid construction
[0110] The nucleic acid sequence of RFP is shown in SEQ ID NO. 7,
[0111] The protein sequence of RFP is shown in SEQ ID NO.8,
[0112] The primers used to amplify the RFP gene are as follows:
[0113] Upstream primer: RFP-F: 5'CGGAATTCGCCACCATGGCCTCCTCCGAGGACGT 3'
[0114] Downstream primer: RFP-R: 5'TCGAGCTCGTTAGGCGCCGGTGGAGTGG 3'
[0115] The 5 end of the upstream primer has an EcoR I restriction site, and the underlined GCCACC sequence is used to promote the transcription of the DNA into RNA, and the translation initiation factor binds to the RNA, and this sequence is the initiation codon of the expressed target gene ATG is next to it.
[0116] The 5 end of the downstream primer has a Sal I restriction site.
[0117] According to the method of Example 1, the RFP gene was amplified by PCR, molecular cloning, ligated to pGEM-T easy vector, and then transcribed in vitro, and ...
Embodiment 3
[0133] Yellow fluorescent protein (YFP) expressed in Aspergillus niger cells:
[0134] 1. Plasmid construction
[0135] The nucleic acid sequence of YFP is shown in SEQ ID NO. 9,
[0136] The protein sequence of YFP is shown in SEQ ID NO.10,
[0137] Using the same primers as the GFP in Example 1, and following the same experimental steps and methods, the GFP gene was amplified by PCR for molecular cloning, ligated to pGEM-T easy vector, and transcribed in vitro, and the resulting RNA was transformed Host spores.
[0138] 2. Use yellow fluorescent protein coding RNA to express yellow fluorescent protein in resting spores of Aspergillus niger, the steps are as follows:
[0139] 1) Aspergillus niger culture and spore collection
[0140] In a 15cm petri dish, prepare a solid agar medium (PDA medium), inoculate the surface of the solid agar medium with Aspergillus niger CBS 513.88, incubate for 3 days at a temperature of 40°C and a humidity of 60-85%, and let the surface of the medium grow ...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com