Rapid extraction method of black sea bream genome DNA
An extraction method and genome technology, applied in the field of rapid extraction of black sea bream genomic DNA, can solve the problems of inapplicable genomic DNA extraction, heavy workload of genomic DNA extraction, and injury to experimental operators, achieving good application prospects and convenient extraction , The effect of low reagent cost
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0025] DNA was extracted from ethanol-preserved black sea bream dorsal fin ray samples and used for PCR amplification of mitochondrial COI gene. The implementation steps are as follows:
[0026] (1) Cut 20 μg of absolute ethanol and soak the black sea bream fin ray sample in a 1.5mL centrifuge tube, and fully grind the sample on the tube wall with a glass grinding rod;
[0027] (2) Add 200 μL of extraction buffer (1% TritonX-100+100mM Tris-HCl (pH=8.0)+300mMNaAc+500mM NaCl), shake for 1min and mix thoroughly;
[0028] (3) Place the metal bath or constant temperature water bath at 65°C for 30 minutes, and mix it occasionally;
[0029] (4) Take out the sample, cool it, and centrifuge it at 13000r / min for 3min in a centrifuge;
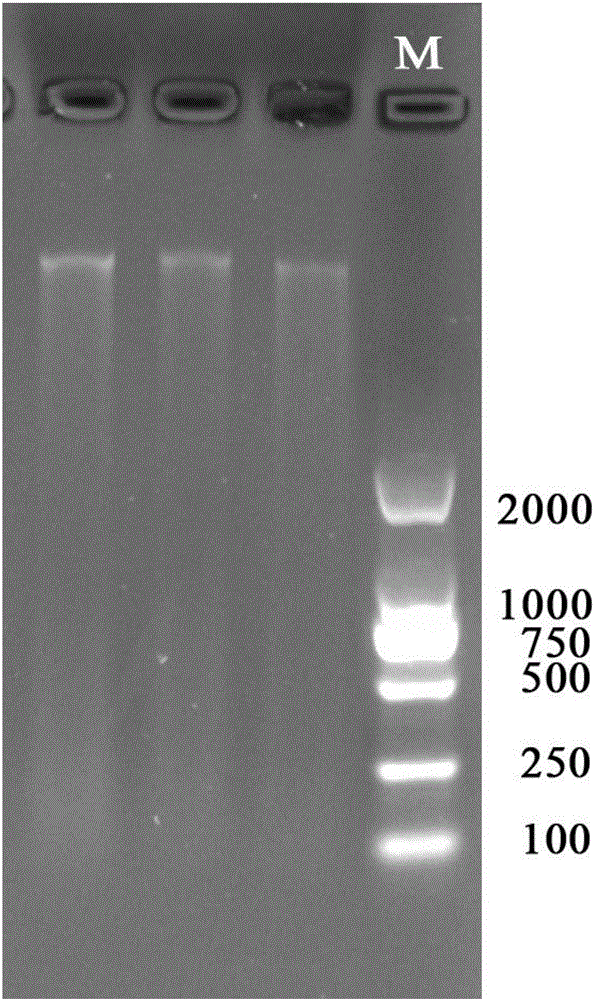
[0030] (5) Aspirate the supernatant into another new tube for the next step of PCR amplification. The black sea bream genome DNA extracted by the method is relatively complete and the effect is ideal. The agarose gel electrophoresis picture of the bla...
Embodiment 2
[0032] (1) Cut 20 μg of black sea bream fresh muscle sample into a 1.5mL centrifuge tube, and fully grind the sample on the tube wall with a glass grinding rod;
[0033] (2) Add 300 μL of extraction buffer (1% TritonX-100+100mM Tris-HCl (pH=8.0)+300mMNaAc+500mM NaCl), shake for 1min and mix well to obtain black sea bream tissue extract;
[0034] (3) Place the constant temperature water bath at 65°C for 35 minutes, and mix it occasionally;
[0035] (4) Take out the sample, cool it, and let it stand for 5 minutes;
[0036] (5) The supernatant is the obtained black sea bream genomic DNA, which is used for the next step of PCR amplification.
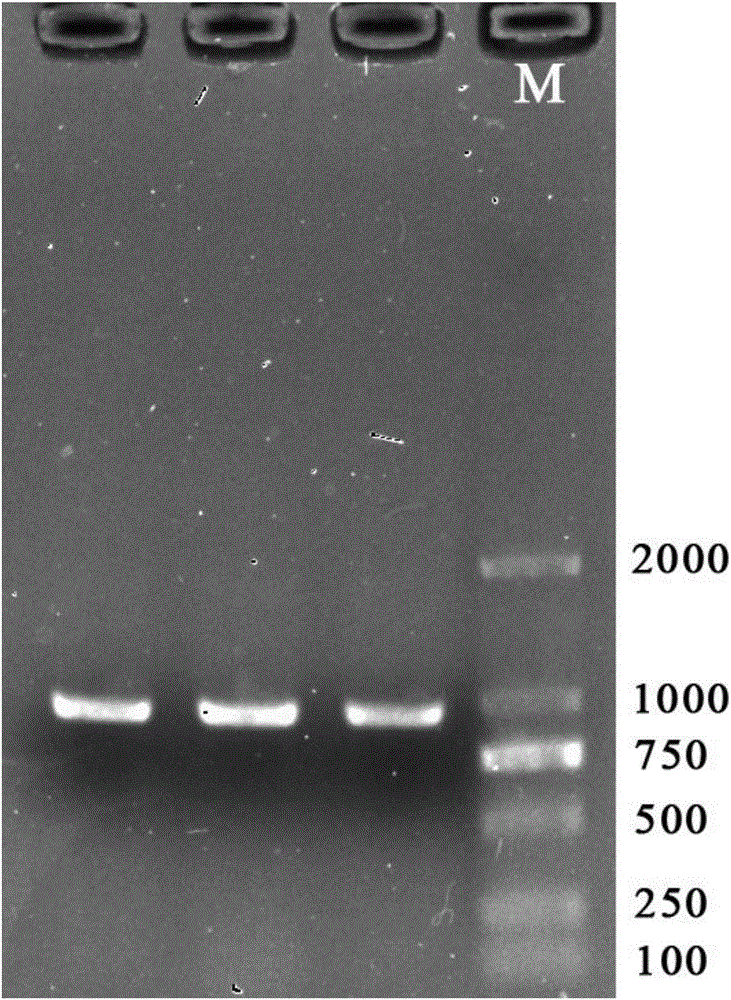
[0037] (6) Use a pair of black sea bream mitochondrial COI gene primers to carry out PCR amplification on the above-mentioned extracted DNA, and the primer sequence is:
[0038] Upstream primer: 5′-TACTCCTTACAGACCGAAAT-3′;
[0039] Downstream primer: 5′-TTATGAAGAAAGTGGCAGA-3′;
[0040] Use 25μL amplification system: 2.5μL 10×PCR buffer, ...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com