Detection primers of acinetobacter lwoffii and fluorescent quantitative PCR detection method
A technology of Acinetobacter lwedenii and fluorescence quantification, applied in the field of bioengineering, can solve the problems of cumbersome medium separation and identification method, deviation of food contamination degree, false negative, etc., to achieve short detection time, ensure food safety, and specificity. strong effect
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0047] 1. Design and synthesis of primers:
[0048] By comparing the genome sequences of Acinetobacter ruckeri and its related species, such as Acinetobacter baumannii, Acinetobacter schindleri, Acinetobacter haemolyticus, etc., to find out The interspecies-specific sequence of Acinetobacter ruckeri, and then BLASTed the sequence to analyze its homology with non-Acinetobacter microorganisms, and finally determined that the β-lactamase gene of the OXA134 family was not Specific conserved gene sequences of A. Based on this gene design specific primers for analysis.
[0049] When designing primers, firstly, according to the β-lactamase amino acid sequence of A.lwoffii ATCC1530 (GenBank: WP_004728961), compare various Acinetobacter ruckerii in the database, A.lwoffii strain St17095 (GenBank: AHA11126.1), A.lwoffii AL3 (GenBank: ADM47435.1) and A.lwoffii strain S459 (GenBank: ALM26450.1), find out the conserved sequence segment between species; then perform DNAMAN software homolo...
Embodiment 2
[0063] In Example 2, the real-time fluorescent quantitative PCR method for detecting Acinetobacter ruckeri provided by the present invention is used to detect raw milk sampled from 8 dairy farms in Yangzhou City, Jiangsu Province in different months (November, December, January, February) , to determine whether the raw milk of each pasture is contaminated with Acinetobacter ruckeri (A.lwoffii), so as to monitor the quality of milk source of each pasture.
[0064] The specific method is as follows:
[0065] (1) Extract the DNA in the raw milk sample to be tested:
[0066] Take 1mL of raw milk, centrifuge at 12000g / min for 5min, discard the upper layer of fat, wash twice with sterile saline, centrifuge at 12000g / min for 2min, and then use the patent "method for extracting total DNA from milk" (patent application number: 200810115822.9) The method for extracting the DNA in the lower layer of the pellet.
[0067] (2) using the DNA extracted in step (1) as a template, amplifying ...
Embodiment 3
[0073] In Example 3, the sensitivity of the fluorescent quantitative PCR detection method of the present invention and conventional PCR was compared.
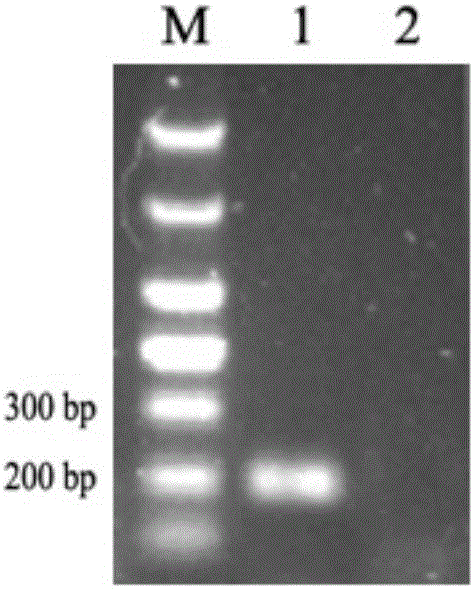
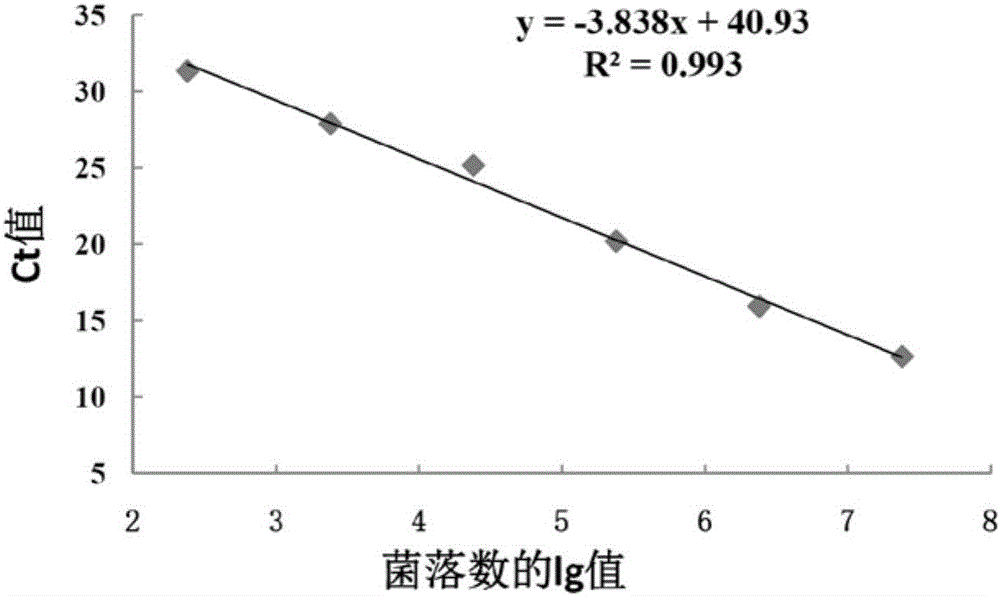
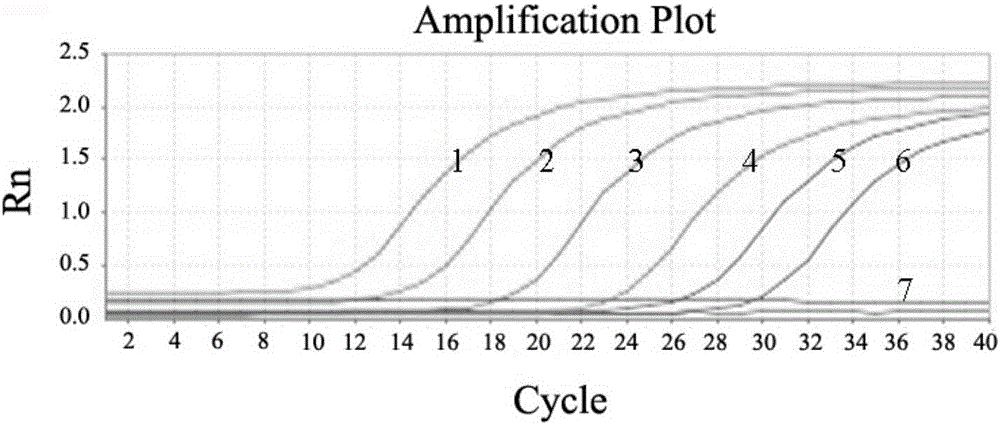
[0074] With the genomic DNA of different concentrations of bacterial liquid obtained in step 4) of Example 1 as a template, AL-F and AL-R are primers for conventional PCR amplification. The reaction conditions are: 95°C for 3min, 30 cycles (95°C 30s, 55°C 30s, 72°C 30s), 72°C 10min. Depend on Figure 4 It can be seen that the sensitivity of conventional PCR detection of A.lwoffii is 2.4×10 3 cfu / mL; compare it with image 3 The minimum number of colonies (2.4 × 10) detected by the fluorescent quantitative PCR of the present invention shown 2 cfu / mL), the sensitivity of the present invention is significantly higher than that of ordinary PCR detection.
PUM
| Property | Measurement | Unit |
|---|---|---|
| Sensitivity | aaaaa | aaaaa |
Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com