Application and inhibitory preparation of siRNA of long non-coding RNA LINC00152
A long-chain non-coding and reagent technology, applied in the application field of siRNA, can solve the problems of unknown function of lncRNA
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
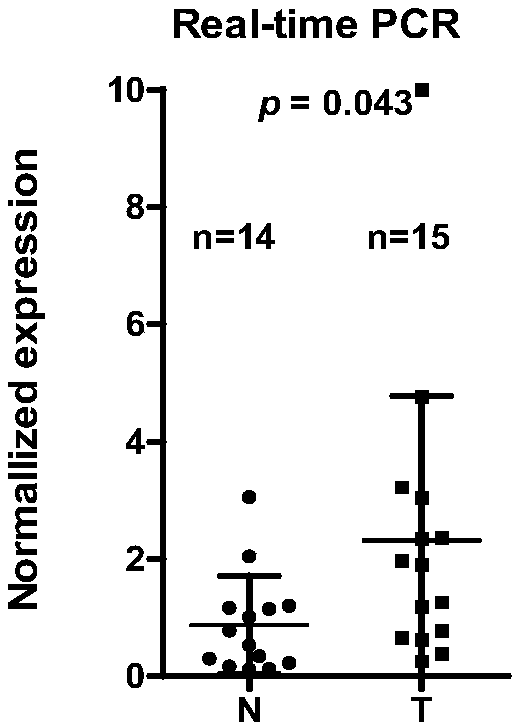
[0030] Example 1, real-time fluorescent quantitative detection confirmed the upregulation of LINC00152 in oral tumors
[0031] 1. Materials and methods:
[0032] Total RNA was extracted from 14 cases of normal oral mucosa and 15 cases of oral tumor tissues, and 2 μg of RNA was reverse-transcribed into cDNA, followed by real-time fluorescent quantitative PCR.
[0033] Forward primer: 5'-TTGATGGCTTGAACATTTGG-3',
[0034] Reverse primer: 5'-TCGTGATTTTCGGTGTCTGT-3'.
[0035] Internal reference gene GAPDH-specific PCR primers for control:
[0036] Forward primer 5'-ACCACAGTCCATGCCATCAC-3'
[0037] Reverse primer 5'-TCCACCACCCCTGTTGCTGTA-3'.
[0038] Real-time fluorescence quantitative PCR reaction system
[0039]
[0040] Real-time fluorescent quantitative PCR reaction steps
[0041]
[0042] After the reaction, the amplification curve and melting curve of real-time fluorescent quantitative PCR were confirmed, and the expression intensity of each gene was standardized a...
Embodiment 2
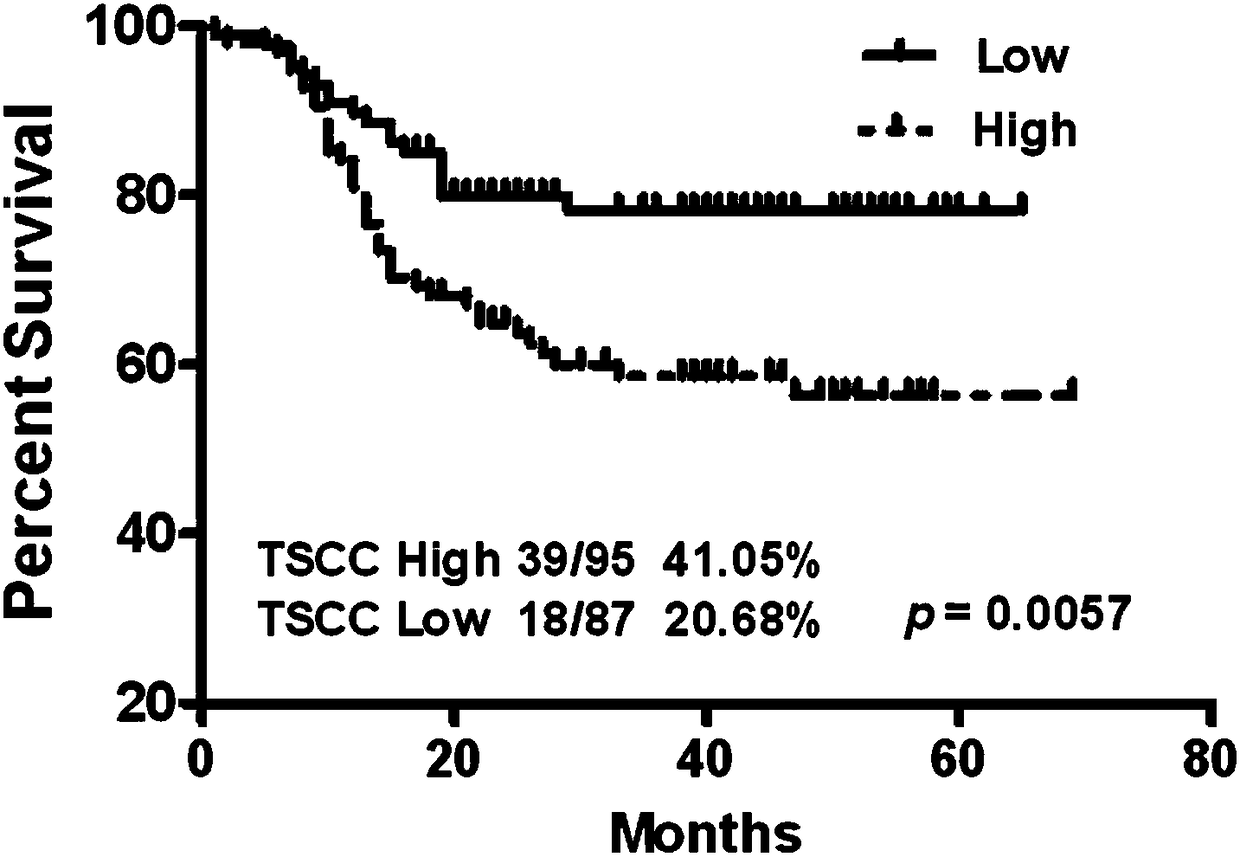
[0045] Example 2, in situ hybridization detection found that the expression of LINC00152 in oral tumors is related to the prognosis of patients
[0046] 1. Material method
[0047] 1.1 Oligonucleotide probes for in situ hybridization of oral tumors and normal control tissues
[0048] Based on the LINC00152 gene sequence analysis, three oligonucleotide probes with the best hybridization parameters and the best specificity were designed. In addition, the housekeeping gene GAPDH was used as a positive control.
[0049] Oligonucleotide probes for in situ hybridization detection of LINC00152 expression:
[0050] LINC00152 Probe 1: 5'-CTATGTCTTAATCCCTTGTCCTTCATTAAAAGC-3';
[0051]LINC00152 Probe 2: 5'-CTTCATTGAACAGTTTGTATATTGGAAACTTGCC-3';
[0052] LINC00152 Probe 3: 5'-GCTGCTTTTAAGTTTCAAATTGACATTCCAGAC-3'.
[0053] Positive control probe (to detect the housekeeping gene GAPDH):
[0054] GAPDH probe 1: 5'-CCACTTTACCAGAGTTAAAAGCAGCCCTGG-3';
[0055] GAPDH probe 2: 5'-GTCAGAGGAG...
Embodiment 3
[0117] Example 3, siRNA interferes with the expression of LINC00152
[0118] 1. Material method
[0119] 1.1 Reagents and kits
[0120] TRIZOL TM Reagent (Invitrogen);
[0121] Reverse transcription kit (#A3500, Promega);
[0122] Antibiotic G418 (Ameresc).
[0123] 1.2 Design of shRNA
[0124] Firstly, input LINC00152 sequence into Invitrogen's Block-It RNAi designer software to find the best siRNA target of this lncRNA, and select the best 2 corresponding target sequences as follows:
[0125] siRNA-1: 5′-CAGGGAAUCUUUCAGCUGGAUUCCG-3′,
[0126] siRNA-2: 5′-UCUAUGUGUCUUAAUCCCUUGUCCU-3′,
[0127] Negative control Scramble sequence: 5'-GACACGCGACUUGUACCAC-3'.
[0128] 1.3 Cell culture and transfection
[0129] The oral cancer cell line Tca8113 was purchased from the Cell Center of Central South University. The RPMI 1640 medium and fetal bovine serum used for cell culture, and the trypsin used for digesting cells were all products of Gibco, USA.
[0130] Oral cancer cel...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com