A breast cancer screening kit
A technology of breast cancer and kits, which is applied in the direction of instruments, biochemical equipment and methods, and analytical materials, and can solve the problems of unsatisfactory screening results
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
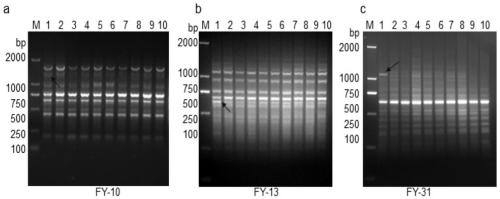
[0034] Example 1 RAPD method specifically amplifies the markers for detecting breast cancer of the present invention (nucleotide sequences shown in SEQ ID NO.1, 2)
[0035] RAPD (Random Amplified Polymorphic DNA)—that is, random amplified polymorphic DNA, is mainly used for molecular detection when the genome information of animals and plants is unknown. RAPD technology relies on PCR, and selects 10 base single-stranded random primers to perform PCR amplification on the DNA of organisms, populations or species genomes to detect differences in genetic loci.
[0036] 1. Experimental method
[0037] (1) Tumor tissue DNA acquisition
[0038] 1. Take 5 pairs of breast cancer tissue and adjacent normal tissue (tissue area 2 cm next to the lesion) (Table 1). Take 5 mg of breast cancer tissue, cut it into pieces, put it in a glass homogenizer, add 2-3ml of nuclei lysis buffer, and homogenize until no tissue lumps are seen;
[0039]2. Transfer all the homogenizer into a 5ml centrifu...
Embodiment 2
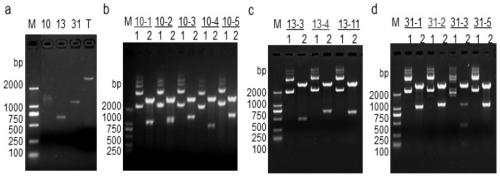
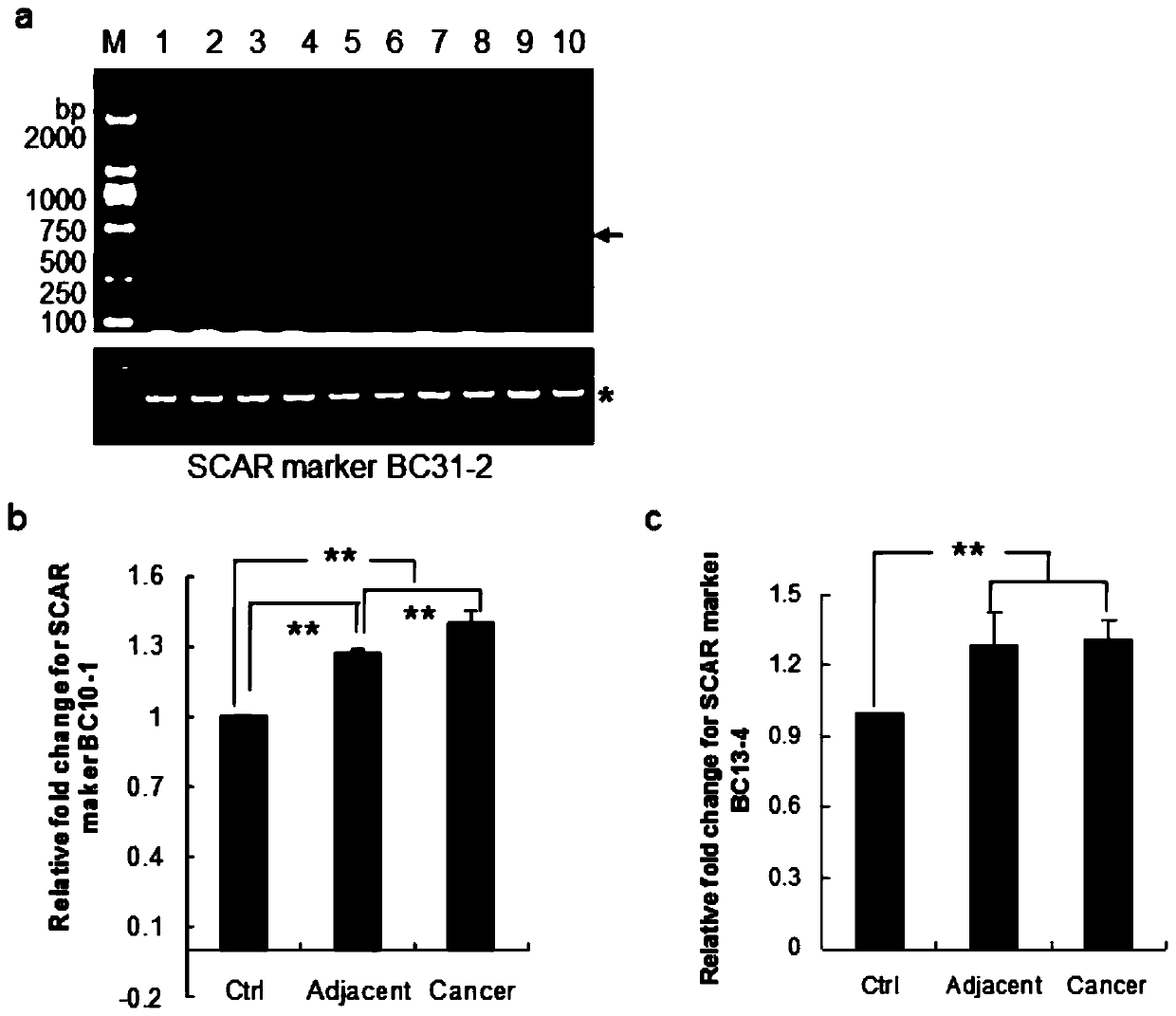
[0153] Example 2 Relationship between expression levels of nucleotide sequences shown in SEQ ID NO.1, 2 and breast cancer (genome level)
[0154] 1. Experimental method
[0155] (1) After sequencing analysis, according to the determined sequence, use Primer3.0 software (http: / / frodo.wi.mit.edu / primer3) to design specific primers and synthesize primers (Table 3). These primers are used to amplify Genomic DNA of tumors and normal people, GAPDH as internal control, they can be used for semi-quantitative PCR.
[0156]
[0157] BC10-1L (SEQ ID NO.3), BC10-1R (SEQ ID NO.4);
[0158] BC31-2L (SEQ ID NO.5), BC31-2R (SEQ ID NO.6);
[0159] GAPDH5G (SEQ ID NO. 15), GAPDH3G (SEQ ID NO. 16).
[0160] At the same time, real-time fluorescent quantitative PCR analysis primers were designed (Table 4). The primers in Table 4 are used to amplify the genomic DNA of tumors and normal people, β-actin is used as an internal reference, and the Taqman probe number (Universal Probe Library, R...
Embodiment 3
[0194] Example 3 Relationship between expression levels of DPEP1 and PHKG2 and breast cancer (mRNA level)
[0195] 1. Experimental method
[0196] (1) Extraction of total RNA from cells and tumor tissues
[0197] Total cellular RNA extraction
[0198] 1. Use miRNeasy Mini Kit or RNeasy Mini Kit purchased from QIAGEN (Cat#217004 or Cat No.74104)
[0199] 2. Take well-grown breast cancer cells with a density of more than 95% (6-well plate), absorb the residual liquid of the medium, and rinse twice with pre-cooled 1×PBS;
[0200] 3. Add 700 μl of QIAzol Lysis (miRNeasy Mini Kit), draw 5 times with a 1ml syringe, and let stand for 5 minutes;
[0201] 4. Add 140 μl of chloroform, shake vigorously for 15 seconds and then let stand at room temperature for 2-3 minutes;
[0202] Centrifuge at 10,000 rpm at 5.4°C for 15 minutes;
[0203] 6. Carefully draw the supernatant into another EP tube, add 1.5 times the volume of absolute ethanol, and turn it upside down several times;
[0...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com