Nucleic acid combination for detecting Her2 gene, kit and application
A kit and gene technology, applied in the field of detection of HER2 gene nucleic acid combinations, can solve the problems of false negatives, cumbersomeness, and expensive probe costs of FISH results, and achieve the effects of reducing false negatives, fast analysis speed, and improving accuracy
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0047] The primers and probes were designed for the human Her2 gene sequence. A total of 3 pairs of primers were designed and synthesized, and a probe was designed correspondingly for each pair of primers. The sequences are shown in Table 3.
[0048] table 3
[0049]
[0050] The size of the fragment amplified by the primer pair of Her2-1 was 155 bp, and the sequence of the amplified fragment was: ACAACCAAGTGAGGCAGGTCGTATTGTTCAGCGGGTCTCCACAACCAAGTGAGGCAGGTCccactgcagaggctgcggattgtgcgaggcacccagctctttgaggacaactatgccctggccgtgctagacaatGGAGACCCGCTGAACAATAC.
[0051] The Her2-2 primer pair amplifies the fragment with a size of 125 bp, and the sequence of the amplified fragment is: TCTTCCTCTCCCTACATCGGGTCCAAGTGAACCAGGGAATCTTCCTCTCCCTACATCGGccccacctgtccccaccccctccagcccacagccatgcccacagccagTTCCCTGGTTCACTTGGAC.
[0052] The size of the fragment amplified by the Her2-3 primer pair was 111 bp, and the sequence of the amplified fragment was: GGGCTCTTTGCAGGTCTCTCCCAGCAAGAGTCCCCATCCTAGGGGCT...
Embodiment 2
[0068] The steps of digital PCR detection are as follows:
[0069] (1) Sample preparation: The source of sample DNA can be blood or neutral formalin-fixed paraffin-embedded tissue sample sections. The DNA sample can be properly diluted to ensure that the sample signal copy number is between 200 copies / μL-2000 copies / μL.
[0070] (2) Prepare the PCR reaction solution according to the following ratio: 2 × PCR master mix (using Life Technologies company's 3D Digital PCR Master Mix V2, catalog number: A26358), primers, probes and sample DNA, made up to a final volume of 15 μL with distilled water to prepare a digital PCR mixture. The content of each primer is 900nM, and the content of each probe is 250nM.
[0071] (3) Brush the prepared 15 μL digital PCR mixture onto the chip.
[0072] (4) Put the chip into a PCR machine, and perform amplification according to the following conditions: pre-denaturation at 95°C for 10 minutes; denaturation at 95°C for 15 seconds, annealing and ...
Embodiment 3
[0075] Screening of primers and probes.
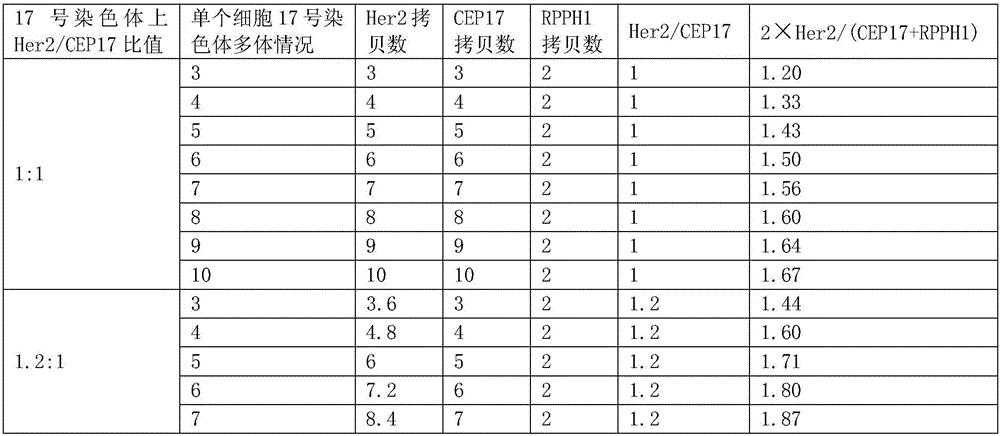
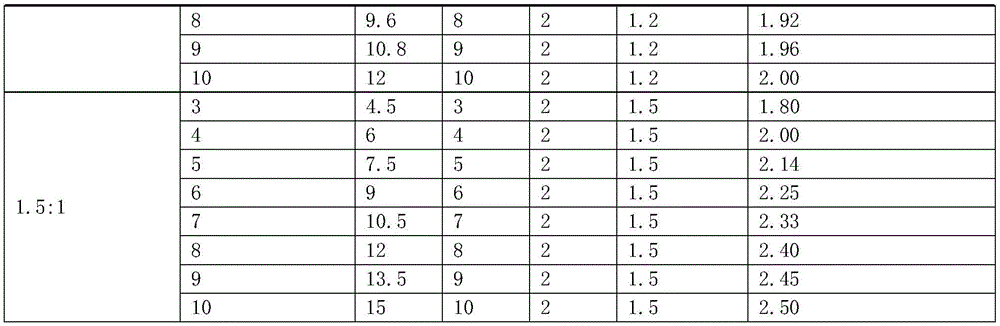
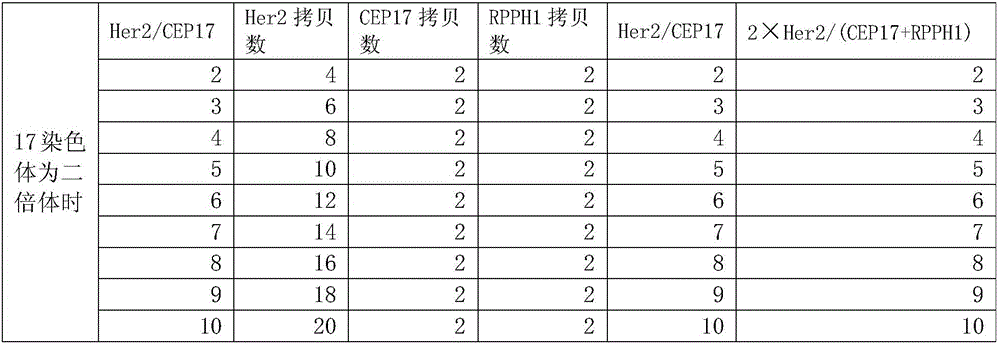
[0076] For genomic DNA, a genome is about 3×10 9 bp, the corresponding molecular mass is 3pg, then the copy number of 1ng genomic DNA is about 333. When doing digital PCR, take 1 ng of normal human blood DNA, make 15 μL of reaction solution, and brush it into the chip. Theoretically, the copy number of the target gene (Her2) and the internal reference gene should be 333, but the digital PCR finally shows FAM (Her2 gene) and VIC (internal reference gene) are expressed in copies / μL, 333 copies / 15 μL≈22 copies / μL. The Her2 gene of normal people is not overexpressed, so the copy number ratio of Her2 to the internal control (FAM / VIC) is theoretically 1:1.
[0077] Screening process:
[0078] Detection is carried out according to the detection method in Example 2.
[0079] Template: 1ng DNA extracted from normal human blood
[0080] Primer probe: the following 15 combinations (respectively Her2-1 / RPPH1, Her2-1 / CEP17, Her2-1 / CEP17-1, Her...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com