Primers for amplifying paphiopedilum type B MADS-box genes
A gene and B-type technology, which is applied in the application of separation of B-type MADS-box genes, genetic relationship analysis, the above-mentioned primers and the kits including the primers, can solve the problem of no cloning and the like
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0068] Example 1. Design of primers for isolating Paphiopedilum class B MADS-box genes
[0069] Download the B gene sequences of other orchids from Genbank, compare the sequences with MEGA 6.0, select the gene conservation region but relatively specific sequence region in Paphiopedilum relatives to design primers. The length of the primer is 18-24bp, and the Tm value is generally controlled at 55-60°C. Avoid using base A at the 3' end of the primer; try not to have a hairpin structure for a single primer, and try not to have a dimer between primers; The GC content of the primer is generally 40-60%, and the GC content of the upstream and downstream primers cannot be too different; the distribution of the four bases in the primer is preferably random, and there is no polypurine and polypyrimidine, especially in the 3' of the primer There should be no more than 3 consecutive G's or C's. Design as many primers as possible.
[0070] Fifteen pairs were selected from the designed...
Embodiment 2
[0071] Example 2. Screening primers for isolating Paphiopedilum class B MADS-box genes
[0072] With reference to the experimental method and materials shown in Example 3 below, the following experiments were performed to screen suitable primer sets:
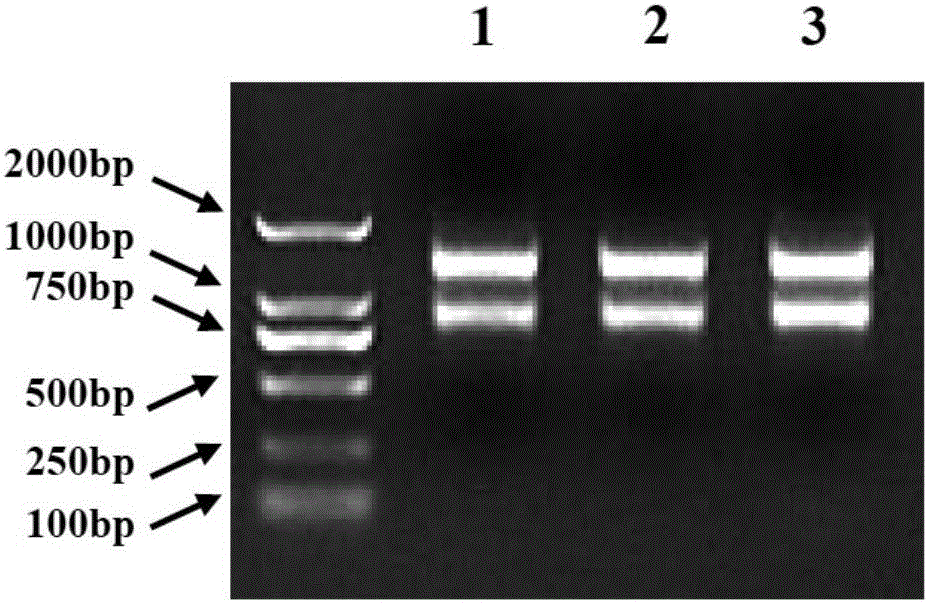
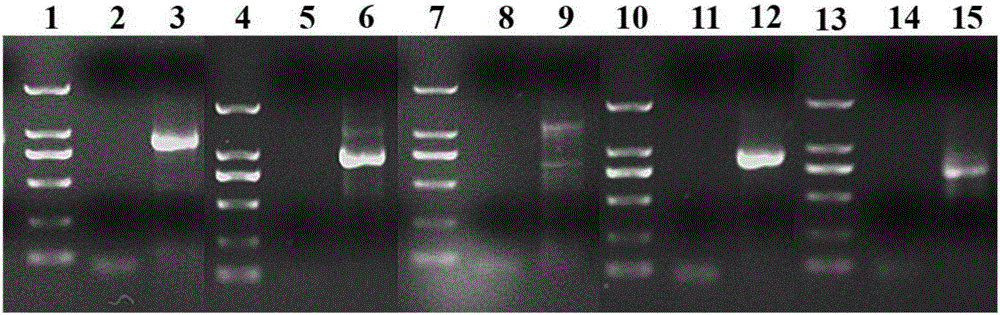
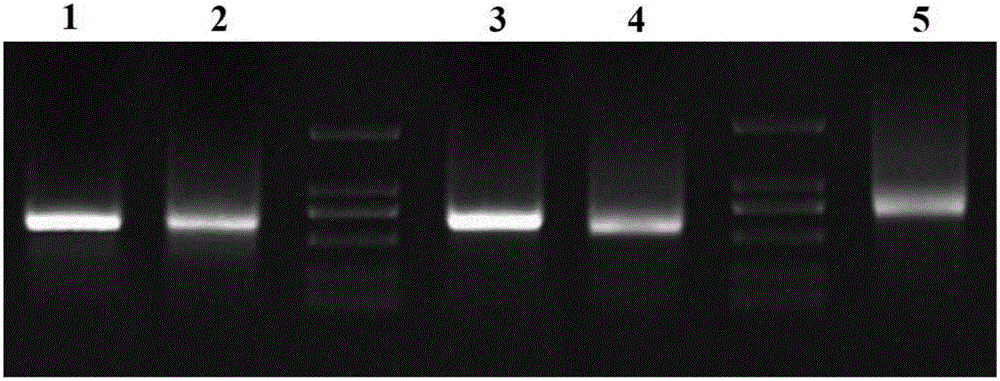
[0073]Utilize existing Oligo(dT) 17 The primers are combined with different F1 primers (AP3A1_F1, AP3A2_F1, AP3B1_F1, AP3B2_F1 or PI_F1) in Table 1-1, and the first round of 3'RACE PCR reaction is carried out. The PCR products are detected by 1% agarose gel electrophoresis, selected and recovered for electrophoresis. Gel DNA with clear and single bands and lengths within the expected range to screen for suitable F1 primers;
[0074] Using Oligo(dT) 17 Combined with different F2 primers (AP3A1_F2, AP3A2_F2, AP3B1_F2, AP3B2_F2 or PI_F2) in Table 1-1, carry out the second round of 3'RACE PCR reaction, detect the PCR product by 1% agarose gel electrophoresis, select and recover the electrophoresis strip Sequence the gel DNA wit...
Embodiment 3
[0080] Example 3. Isolation of Paphiopedilum class B MADS-box gene
[0081] 1. Test material
[0082] (1) Plant material: Henry's Paphiopedilum.
[0083] (2) Cloning vectors: pEASY-T1 and pEASY-T3 (Beijing Quanshijin Biotechnology Co., Ltd.).
[0084] (3) Reagents: RNA extraction was performed using RNA prep Pure Polysaccharide Polyphenol Plant Total RNA Extraction Kit (spin column type, DP441, Beijing Tiangen Biochemical Technology Co., Ltd.); reverse transcription was used to synthesize first-strand cDNA using Invitrogen’s Superscript III reverse transcription kit. Transcriptase and related system (18080-044), purchased from Beijing Huazhong Haiwei Technology Co., Ltd.; 5'RACE uses Invitrogen's Kit (L150202), purchased from Beijing Juhua Tektronix Technology Co., Ltd.; gel recovery using ordinary agarose gel DNA recovery kit (DP209-02), purchased from Beijing Tiangen Biochemical Technology Co., Ltd.; Rnase Inhibitor (2313A), Takara Taq enzyme and dNTP were purchased fr...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com