LAMP primer group, kit and detection method for rapid test of bacterial polymyxin drug resistance gene mcr-1
A polymyxin and drug-resistant gene technology, applied in biochemical equipment and methods, recombinant DNA technology, microbial measurement/inspection, etc., can solve the problems of cumbersome operation process and long time consumption, and achieve high sensitivity and specificity Good, highly specific effect
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0039] Embodiment 1, rapid detection bacterial polymyxin resistance gene mcr-1 The establishment of the kit
[0040] Rapid detection of bacterial polymyxin resistance genes mcr-1 The kit includes the following components: (1) LAMP detection primer set; (2) DNA polymerase; (3) LAMP reaction solution; (4) color reagent; (5) positive control and negative control.
[0041] (1) LAMP detection primer set
[0042] bacterial polymyxin resistance gene mcr-1 (GenBank accession number: KX886345.1), for its 6 regions, LAMP primer design was carried out, after a large number of primer specificity experiments, a LAMP detection primer set with good specificity was selected, including a pair of inner primers and a pair of outer primers. The nucleotide sequences of the primer and a pair of loop primers are as follows:
[0043] Internal primer 1: 5'- GCGATGGGATAGGTTTGGCTGTGTTGCCGTTTTCTTGAC-3' (SEQ ID NO: 1);
[0044] Internal primer 2: 5'-TGCTGACGATCGCTGTCGGCACATAGCGATACGATGAT-3' (SEQ ID NO:...
Embodiment 2
[0054] Embodiment 2, rapid detection bacterial polymyxin resistance gene mcr-1 Methods
[0055] Rapid detection of bacterial polymyxin resistance gene using the kit of Example 1 mcr-1 ,Specific steps are as follows:
[0056] (1) Extract the DNA of the sample to be tested:
[0057] Bacterial genomic DNA was extracted by boiling method.
[0058] (2) Use the LAMP detection primer set to perform isothermal gene amplification reaction on the DNA of the sample to be tested:
[0059] The 25 μL system of isothermal gene amplification reaction is as follows: the final concentration of inner primers 1 and 2 is 8 pmol / µL, the final concentration of outer primers 1 and 2 is 1 pmol / µL, and the final concentration of loop primers 1 and 2 is 4 pmol / µL. µL, 12.5 µL of reaction solution, 8 U of DNA polymerase, 50 ng of DNA of the sample to be tested, make up to 25 µL with sterilized deionized water, and then add 20 µL of sealing solution;
[0060] The isothermal gene amplification program...
Embodiment 3
[0064] Embodiment 3, sensitivity experiment
[0065] Take the positive control substance (that is, the constructed drug containing bacterial polymyxin resistance gene mcr-1 Fragment pMD19-T plasmid), measure its concentration and calculate the copy number, dilute according to 10-fold concentration gradient, select 1.0×10 0 ~1.0×10 9 The concentration of copies / μL was used as a sample for experimentation; the detection method in Example 2 and the conventional PCR method were used for detection respectively.
[0066] The primer sequences used in conventional PCR detection methods are as follows:
[0067] MCR-1 -F:CGGTCAGTCCGTTTGTTC (SEQ ID NO: 7);
[0068] MCR-1 -R: CTTGGTCGGTCTGTAGGG (SEQ ID NO: 8).
[0069] Adopt the detection result of embodiment 2 detection method as figure 2 Shown: its pair mcr-1 The minimum detection limit of the gene is: 1.0×10 4 copies / μL.
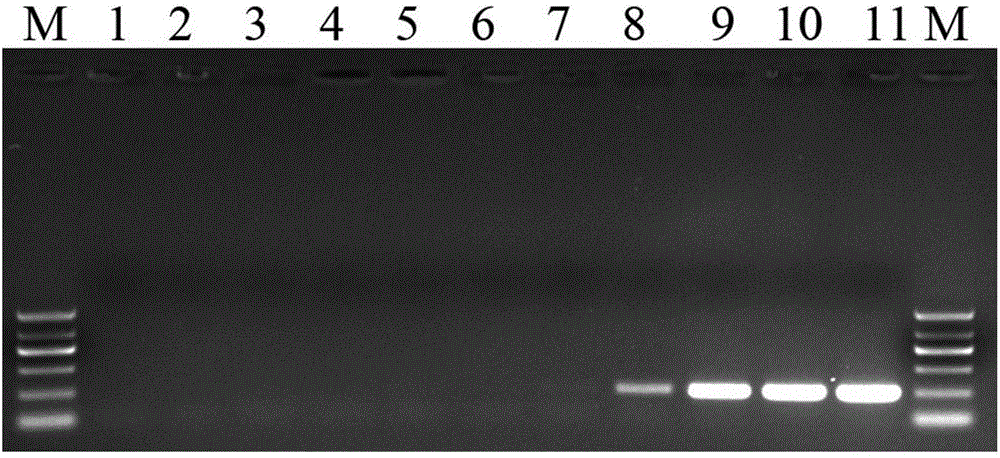
[0070] The detection results by conventional PCR method are as follows: image 3 Shown: its pair mc...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com