DMD (Duchenne muscular dystrophy) gene capture probe and application thereof in DMD gene mutation detection
A gene capture and capture probe technology, which is used in recombinant DNA technology, microbial determination/inspection, DNA/RNA fragments, etc. Inefficiency, etc.
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0063] Embodiment 1, the preparation of DMD gene capture probe
[0064] 1. Sequence design and preparation of single-stranded subprobes
[0065] The sequence of the DMD gene used is chrX:31137345-33357726 of the human reference genome version Hg19 (updated on August 6, 2015). Design the sequence of the DMD gene capture single-stranded sub-probe according to the DMD gene sequence, the length of each single-stranded sub-probe is 60bp, and the sequence of each single-stranded sub-probe is as follows:
[0066] The sequence of the first single-stranded subprobe is the 1-60 position of the DMD gene (ie chrX: 31137345-31137404);
[0067] The sequence of the second single-stranded subprobe is the 58th-117th position of the DMD gene;
[0068] The sequence of the third single-stranded subprobe is the 115th-174th position of the DMD gene;
[0069] ...
[0070] The sequence of the nth single-stranded subprobe is [57(n-1)+1]-[57(n-1)+60] of the DMD gene;
[0071] ...
[0072] The seq...
Embodiment 2
[0087] Embodiment 2, establishment of DMD gene mutation detection method
[0088] 1. Construction of whole genome library of samples to be tested
[0089]1.1 Ultrasound fragmentation: Dilute the whole genome DNA of the sample to be tested with an initial amount of 0.5-3 μg to 30 ng / μL with 1×low TEBuffer (Thermo). Covaris S2 ultrasonic instrument was used for ultrasonic fragmentation, and the value of Covaris system was set according to the standard, 6 cycles × 60s, water bath temperature: 5°C, duty cycle: 20%, intensity: 5, mode: Frequencysweeping, to obtain fragmented DNA.
[0090] 1.2 End filling: Take 100 μL of fragmented DNA from step 1.1, 8 μL of dNTPs, 2 μL EndPolishing enzyme I (10 U / μL, Agilent) and 16 μL End Polishing enzyme II (5 U / μL, Agilent), add water to a total volume of 200 μL After incubation at 25°C for 30 min, the DNA was purified using the PureLink PCR Purification Kit (Invitrogen) to obtain end-filled DNA.
[0091] 1.3 Ligation of P1 and P2 adapters: T...
Embodiment 3
[0118] Embodiment 3, utilize the DMD gene capture probe of embodiment 1 and the DMD gene mutation detection method of embodiment 2 to detect the DMD gene mutation of clinical samples
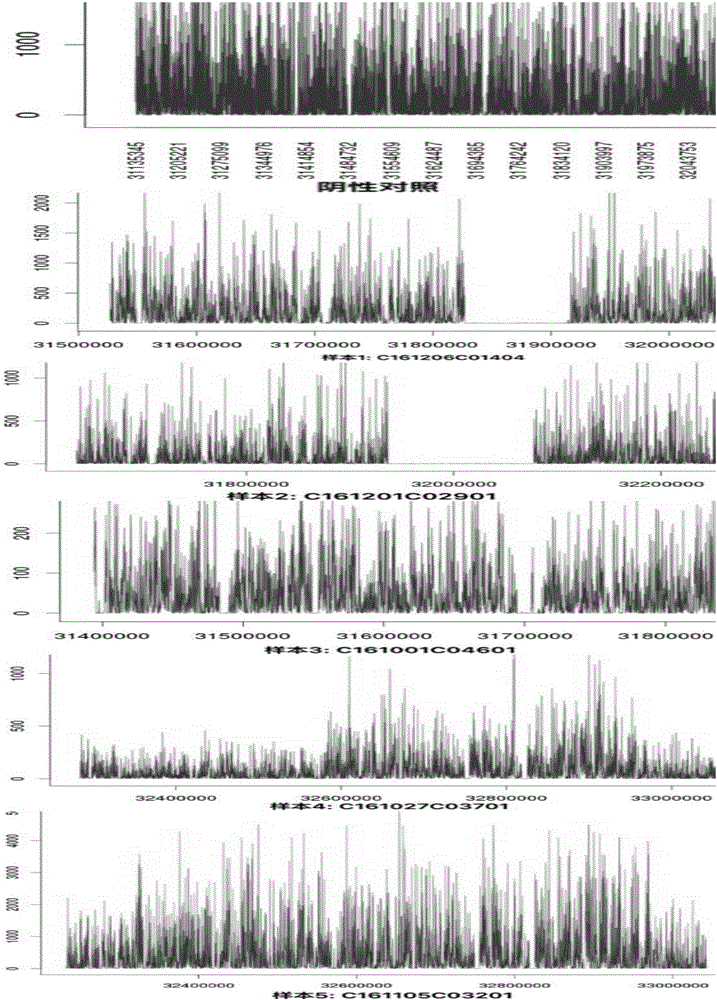
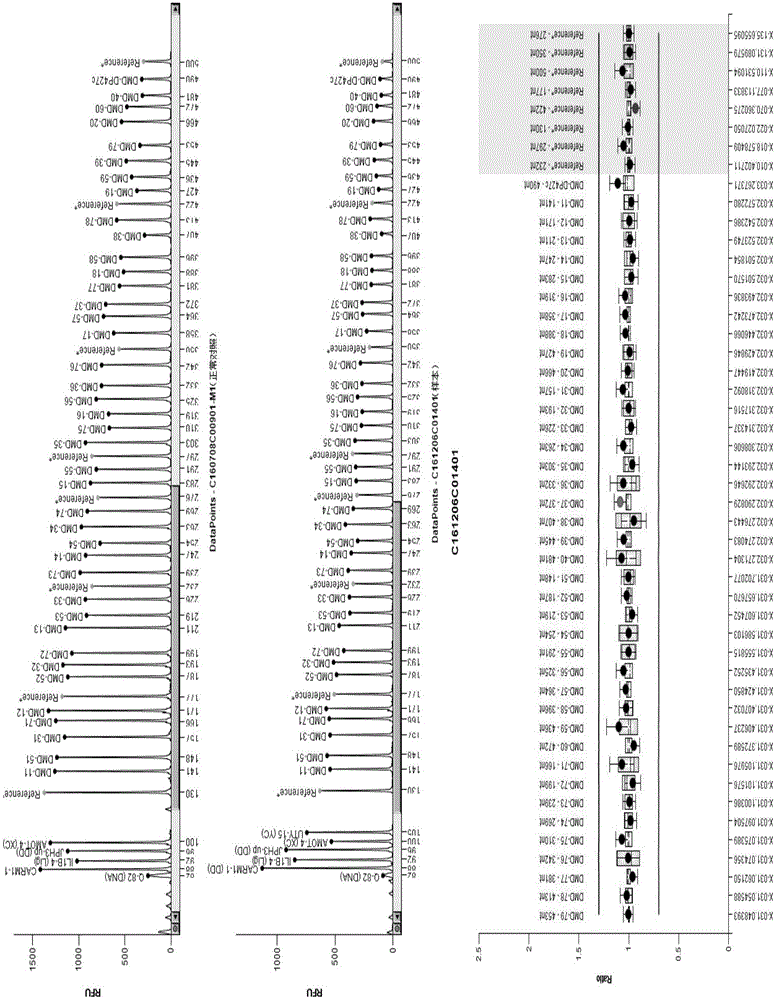
[0119] After informed consent, DNA samples from 5 patients with exon deletion / repeat expansion mutations in the DMD gene were selected (verified by MLPA detection, Figure 2-Figure 11 ) as a sample to be tested, and a DNA sample negative for MLPA detection was used as a normal control (negative control), and the DMD gene mutation detection method of Example 2 was used to detect the DMD gene mutation. The kit used by MLPA to detect whether there is exon deletion / repeat expansion mutation in DMD gene is MLPA P052 detection kit (HRC-Holland).
[0120] The results showed that the high-throughput sequencing images of 5 cases of MLPA-positive samples were shown in figure 1 , see Table 1 and Table 3 for data analysis results. figure 1 In , the negative control refers to the high-throughput sequencing...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com