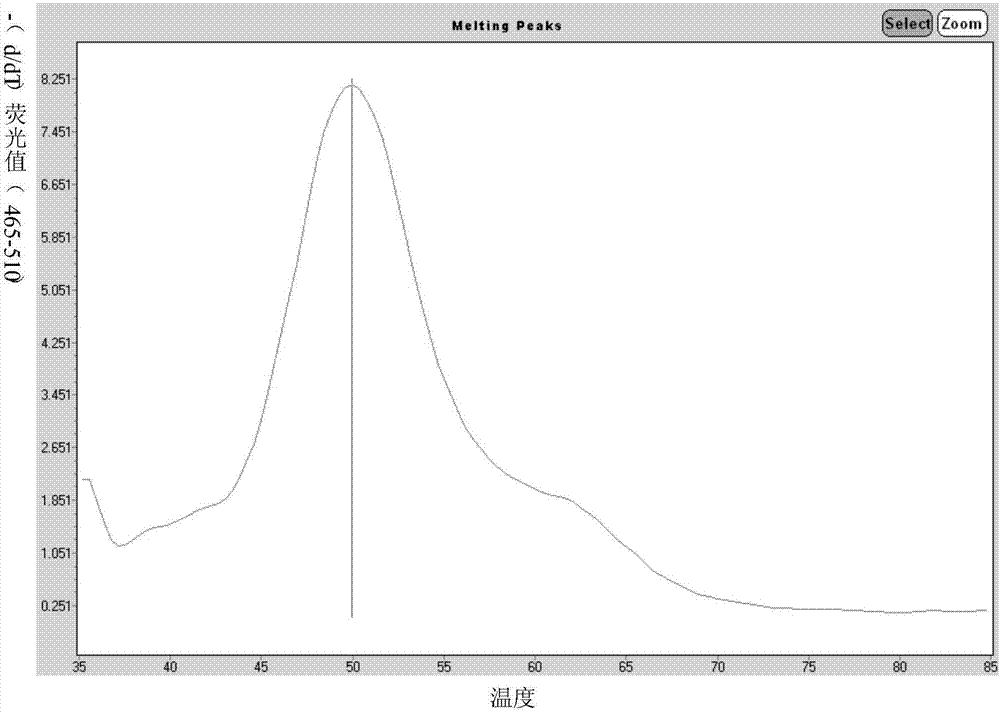
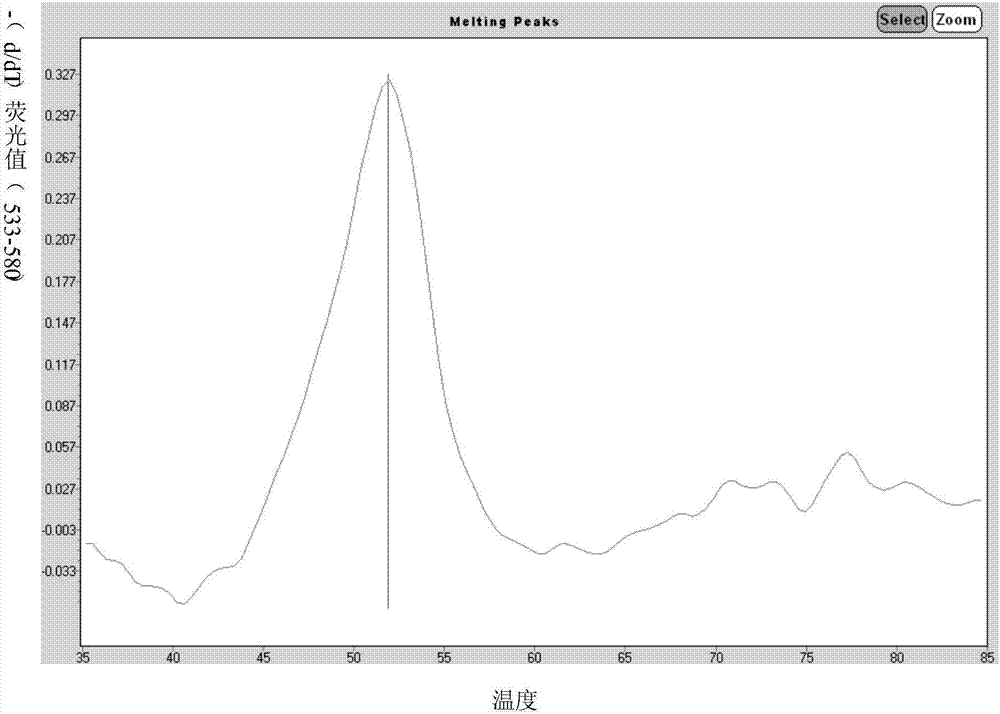
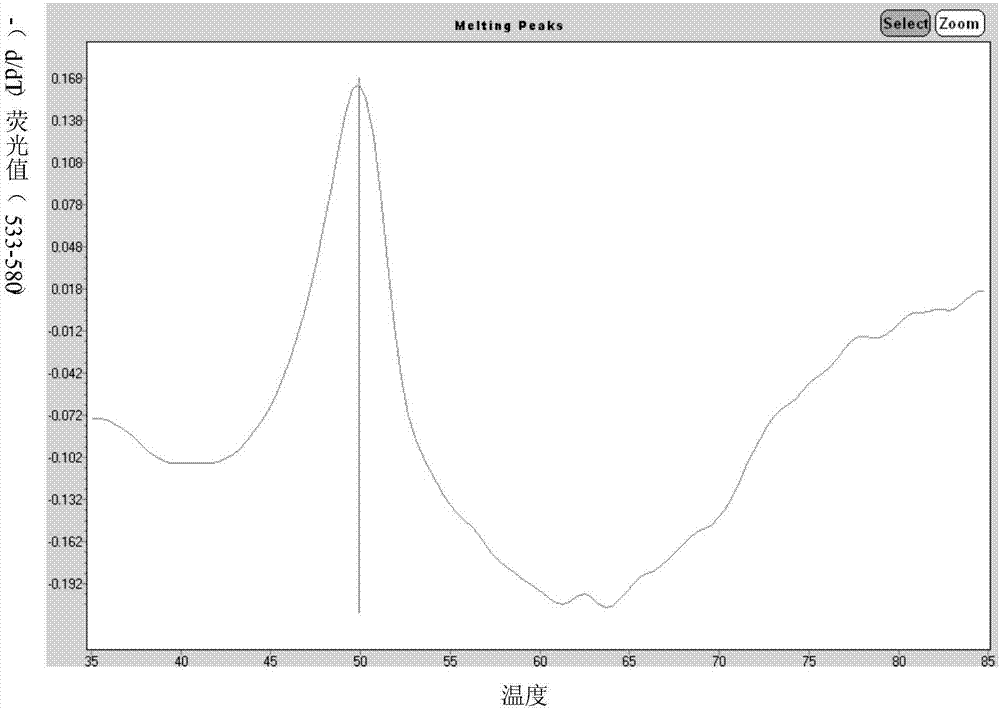
Primers for single-tube simultaneous detection of parainfluenza viruses and coronaviruses based on melting curve assay and application of primers
A technology of coronavirus and melting curve, which is applied in the determination/inspection of microorganisms, resistance to vector-borne diseases, biochemical equipment and methods, etc., to achieve the effect of convenient operation, guaranteed accuracy and good stability
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0061] Example 1: Simultaneously detect the primers and probe sequences of parainfluenza virus types 1 to 3, coronavirus 229E, NL63, OC43, HKU1, synthesized by Yingwei Jieji (Shanghai) Trading Co., Ltd.:
[0062] Reverse transcription primers for reverse transcription:
[0063] Numbering Primer name Sequence (5'-3') corresponding virus SEQ ID NO.1 PIV1-rtp TCTGCACATCCTTGAGTGATTA Parainfluenza virus type 1 SEQ ID NO.2 PIV2-rtp ATCTTCTTTYTCAGARCTTGTAGC Parainfluenza virus type 2 SEQ ID NO.3 PIV3-rtp GCAGTCTCTCTGYGTTTYCC Parainfluenza virus type 3 SEQ ID NO.4 CV229E-rtp GCTAAGGTTGTAKTCACCAAAACT Coronavirus CV229 SEQ ID NO.5 CVNL63-rtp CAAAAGTAGTGTTTATACCAAACTT Coronavirus NL63 SEQ ID NO.6 CVOC43-rtp AAGTTTTGATTACTAACTCCTGGT Coronavirus OC43 SEQ ID NO.7 CVHKU1-rtp AAAAGCAKTAGCTATYAACTTTTGAT Coronavirus HKU1
[0064] The above 7 reverse transcription primers are used together in one dete...
Embodiment 2
[0073] Embodiment 2: sample nucleic acid extraction
[0074] (1) Collection, storage and transportation of clinical specimens: suitable for collection of throat swab specimens. It is advisable to collect in the early morning. Rinse mouth with clean water, assisted by the examiner with a tongue depressor, pass the throat swab over the base of the tongue, reach the lesion in the pharyngeal isthmus, apply it several times, avoid touching the tongue and oral mucosa when taking it out, and swab the swab after sampling Put the swab into about 1mL of normal saline and wash it thoroughly, then squeeze it dry, and discard the cotton swab. The specimens to be tested should not be stored for more than 24 hours at 4°C, and should not be stored for more than 48 hours at -20°C; and not more than three months at -80°C. Specimens should be transported in curlers or foam boxes with ice.
[0075] (2) Extraction of viral RNA: Extraction of viral RNA was carried out according to the instructio...
Embodiment 3
[0076] Example 3: Reverse transcription using the extracted nucleic acid as a template
[0077] (1) Prepare mixed reverse transcription primer working solution:
[0078] Dilute the synthesized reverse transcription primers to 100uM, add 10uL each to a 1.5mL centrifuge tube, add water to 500uL, vortex and mix to obtain the reverse transcription primer working solution.
[0079] (2) Sample viral RNA reverse transcription:
[0080] The total RNA in the sample was reverse-transcribed according to the RevertAid RT Reverse Transcription Kit (Themo, K1691). The reaction system was 10uL, the reverse transcription primer working solution was 0.5ul, other components were added according to the instructions, and finally water was added to 10uL.
[0081] Reaction procedure: 1h at 42°C; 5min at 85°C; store at 4°C.
PUM
| Property | Measurement | Unit |
|---|---|---|
| melting point | aaaaa | aaaaa |
Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com