Method of simultaneously performing enzyme-cut and link up co-system based on CcdB lethal gene and SmaI restriction enzyme cutting site
A technology of enzyme cutting sites and genes, applied in the fields of biochemistry and molecular biology, to achieve the effect of speeding up the experiment progress, broad application prospects, and reducing experiment costs
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0024] 1 Materials and methods
[0025] 1.1 Materials, reagents and strains
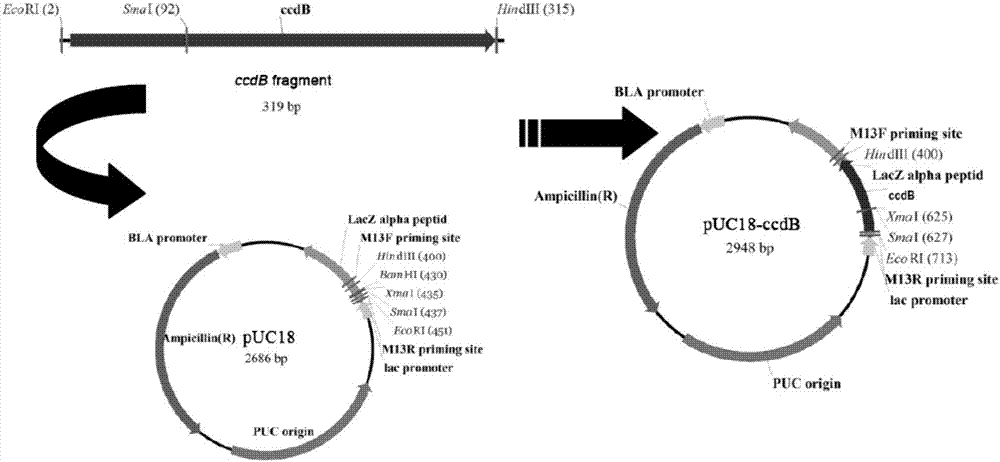
[0026] The Reading Frame CassetteB plasmid containing the ccdB lethal gene (its Genbank accession number is EU496090.1) was purchased from Invitrogen, the restriction enzymes EcoRI, HindIII, and SmaI were purchased from NEB, and the high-fidelity DNA polymerase Phusion-HF, T4 DNA The ligase was purchased from Thermo Fisher Scientific, and the backbone vector was pUC18. Escherichia coli strains E.coli DH 5α and DB 3.1 were preserved by our laboratory. Primers were synthesized by Huada Biological Company.
[0027] 1.2 Method
[0028] 1.2.1 Amplification of ccdB gene
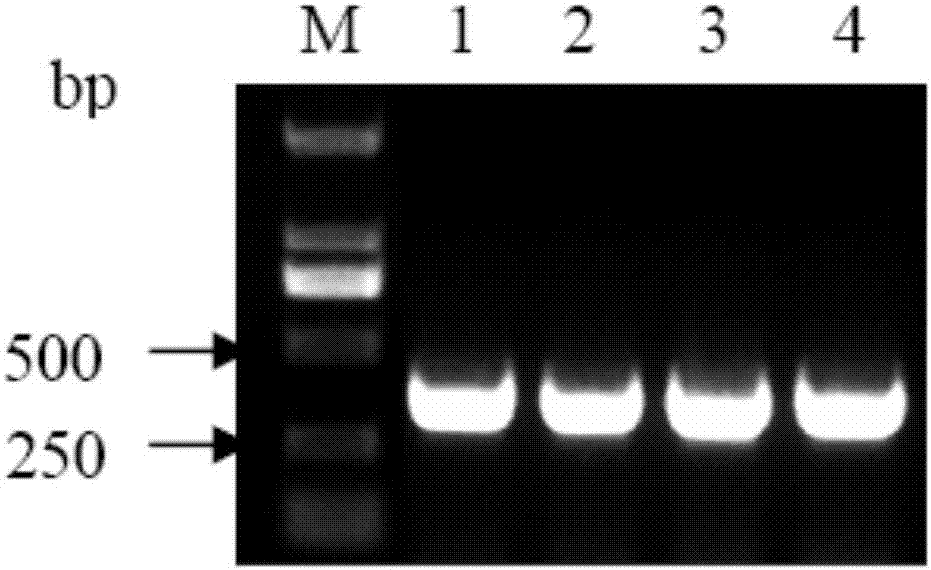
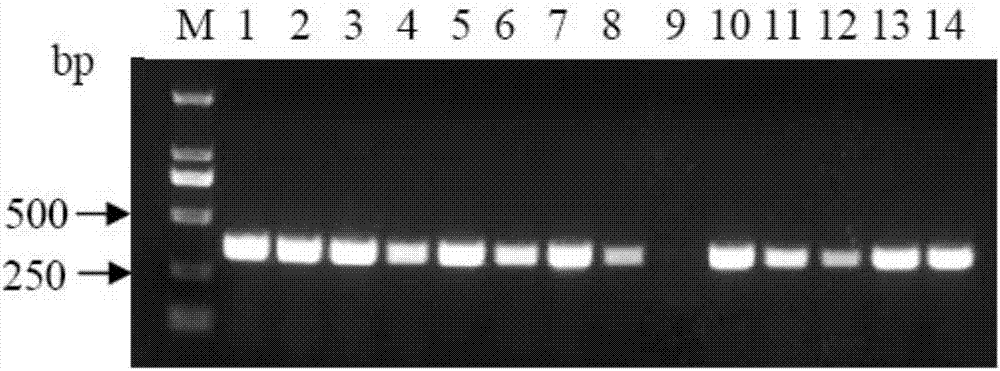
[0029] Using Reading Frame Cassette B as a template, use Phusion DNA polymerase and the primers ccdB-F and ccdB-R in Table 1 to amplify the ccdB lethal fragment. The amplification program is: pre-denaturation at 98°C for 30s, denaturation at 98°C for 10s, annealing at 56°C for 30s , 72°C extension for 15s, 30 cycles, the final extens...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com