TaqMan-MGB fluorescent quantitative polymerase chain reaction (PCR) detection primer, TaqMan-MGB fluorescent quantitative PCR detection probe and TaqMan-MGB fluorescent quantitative PCR detection method for seneca valley virus (SVV)
A technology of fluorescence quantification and detection method, which is applied in biochemical equipment and methods, microbiological determination/inspection, DNA/RNA fragments, etc. It can solve the problems of inability to do accurate quantitative analysis, death of young piglets, insufficient resolution of samples, etc. problems, to achieve the effect of facilitating probe design, shortening the length of the probe, and shortening the length of the amplified fragment
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0041] 1. Primer and TaqMan-MGB probe design
[0042] Retrieve and obtain the whole genome sequence of Seneca Valley virus from GenBank (GenBank accession number is NC_011349, KC667560 and KT321458), carry out sequence alignment by DNAstar software, find out the conserved sequence of Seneca Valley virus genome specificity, then BLAST further Determine its conservatism.
[0043] Primers were designed for this conserved sequence by Oligo 6.0 primer design software to obtain a pair of specific primers and a MGB probe. The primers and probe were located near the 3' end of a single ORF (7021-7089 positions). 69bp.
[0044] Upstream primer: 5'-TTATAGAATTTGGAAGCCATGCT-3' (SEQ ID NO:1)
[0045] Downstream primer: 5'-ATCAGAATGCAGCTTCTCGAGTAG-3' (SEQ ID NO:2)
[0046]Probe: FAM-5'-CCTACTTCAAACCAGGAAC-3'-MGB-NFQ (SEQ ID NO:3)
[0047] 2. Construction and preparation of quantitative standard plasmids
[0048] On the genome sequence on both sides of the above primers (SEQ ID NO:1 and ...
Embodiment 2
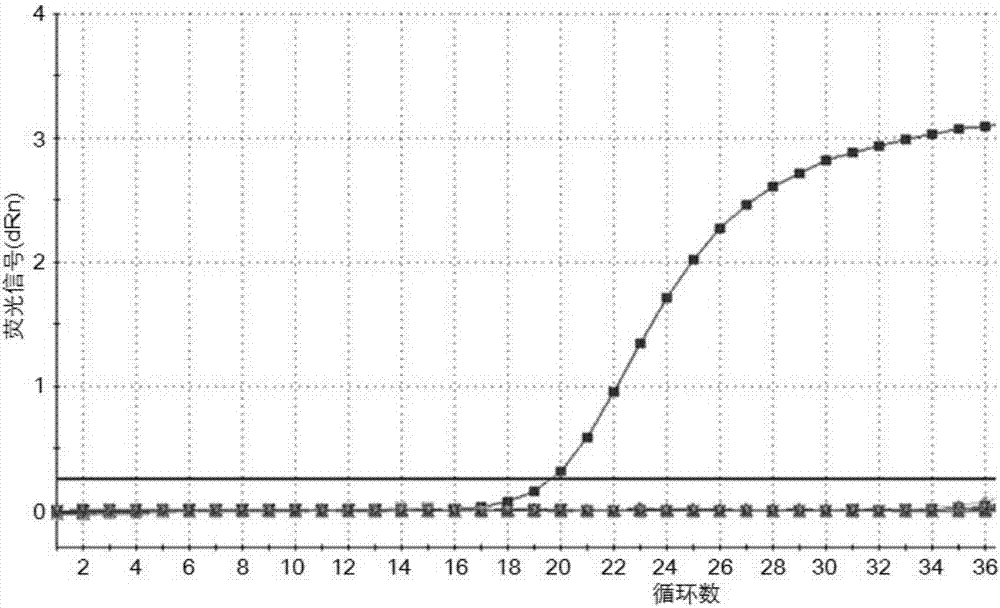
[0064] Example 2 Seneca Valley virus TaqMan-MGB fluorescence quantitative PCR method specificity experiment
[0065] SVV-positive samples, PRV-positive samples, PCV2-positive samples, PRRSV-positive samples, FMDV-positive samples, PEDV-positive samples, RV-positive samples, CSFV-positive samples, PDCoV-positive samples were respectively used as templates, and distilled water was used as a negative control according to Example 1. The operation method of TaqMan-MGB fluorescent quantitative PCR reaction is carried out separately, and the reaction results of each sample are observed and recorded in real time, such as figure 2 As shown, only SVV-positive samples showed a good S-shaped curve, and the remaining samples did not show amplification curves, indicating that the method has good specificity.
Embodiment 3
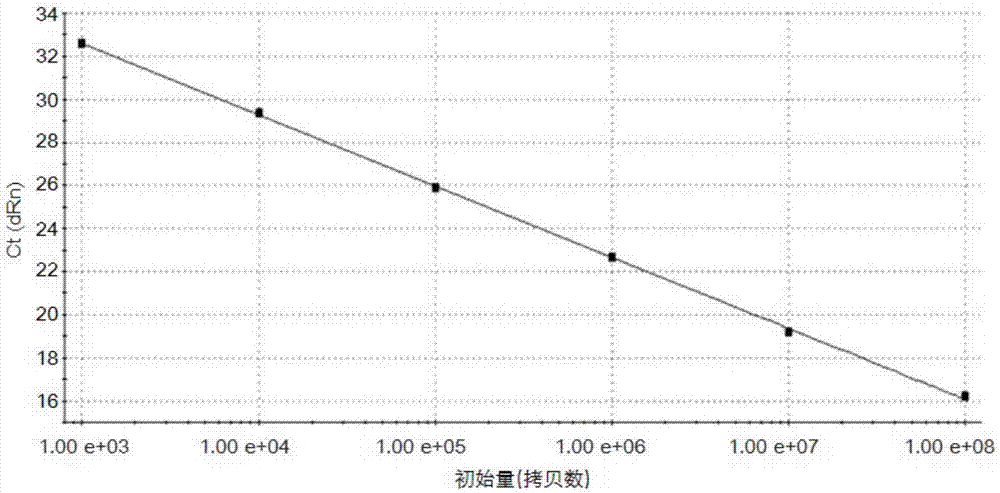
[0066] Example 3 Sensitivity experiment of Seneca Valley virus TaqMan-MGB fluorescence quantitative PCR method
[0067] With the concentration gradient prepared in embodiment one being 10 1 ~10 8 The standard plasmid sample of copies / μL is used as a template, and the reaction is carried out respectively according to the operation method of the TaqMan-MGB fluorescent quantitative PCR reaction in Example 1, and the reaction results of each sample are observed and recorded in real time, such as image 3 As shown, all concentrations of samples showed good amplification curves, indicating that the method has good sensitivity.
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com