Subunit coronavirus vaccine for dimerization-based receptor binding domains
A coronavirus, purpose technology, applied in the field of medicine, can solve the problems of limited immunogenicity and insufficient safety evaluation of fusion proteins
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0032] Example 1: Preparation of recombinant baculoviruses expressing MERS and SARS antigens
[0033]After the MERS-CoV RBD nucleotide sequence (as shown in SEQ ID NO: 3) and the SARS-CoV RBD (as shown in SEQ ID NO: 4) nucleotide sequence 3' end are added with a translation stop codon, they are cloned into a Between the EcoR I and Xho I restriction sites of the pFastBac vector (pFastBac-SP) of the gp67 signal peptide, the protein coding region is fused and expressed behind the signal peptide gp67 sequence for the secretion of the target protein. The vectors pFastBac-SP-MERS-RBD and pFastBac-SP-SARS-RBD were obtained respectively.
[0034] The vector used to express the MERS monomeric RBD is inserted into the EcoRI and XhoI restriction sites of pFastBac-SP by inserting the nucleic acid fragment from amino acid 379 to 589 in the S protein sequence (GenBank: AFS88936.1 on NCBI), and The 3' end is composed of 6 histidines. The vector used to express the SARS monomer RBD is mutat...
Embodiment 2
[0036] Embodiment 2: Purification of MERS-RBD and SARS-RBD
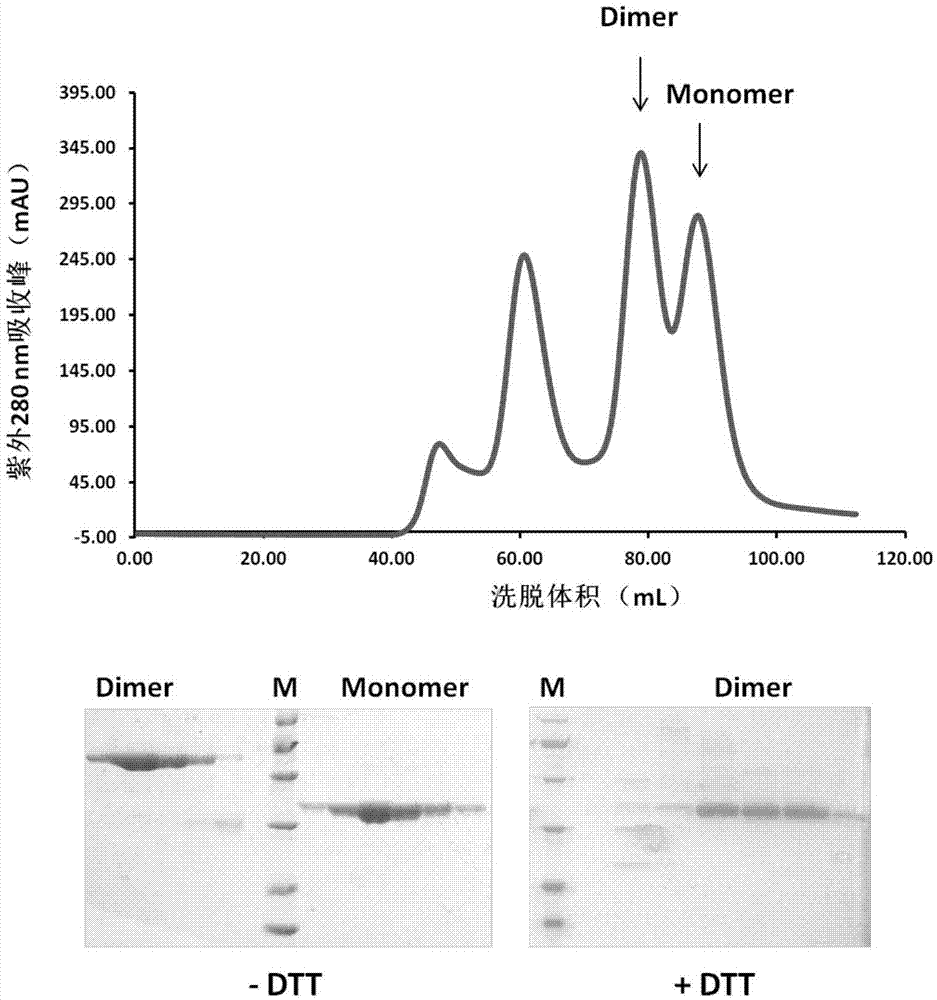
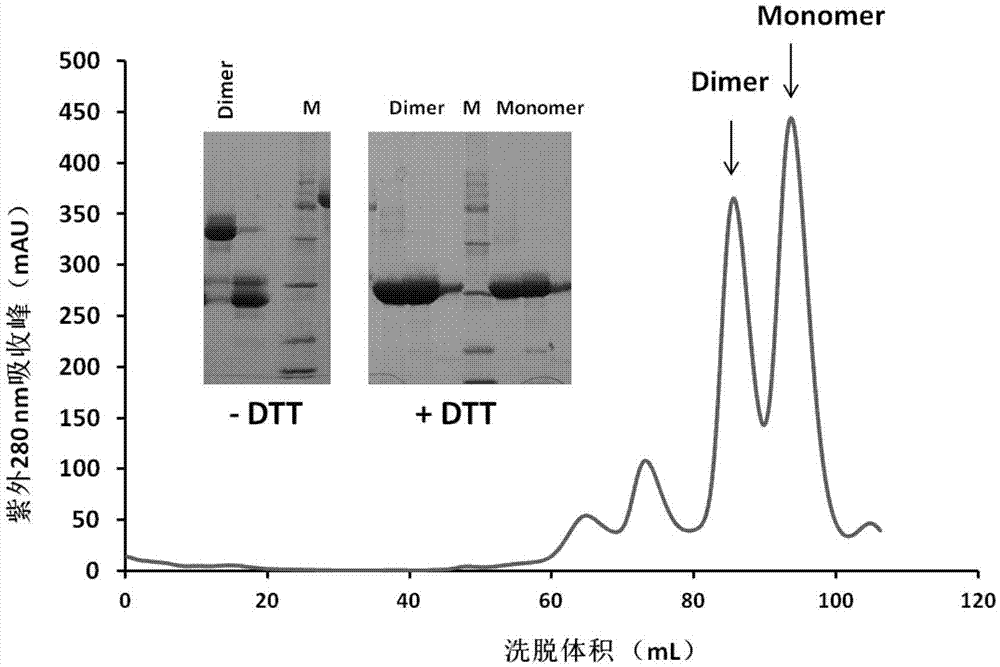
[0037] Filter the supernatant of cells expressing MERS-RBD and SARS-RBD through a 0.22 μm filter to remove cell debris. The target protein was captured by a HiTrap QHP anion exchange column, and then crudely purified by elution with a buffer containing 1M NaCl. After identification by SDS-PAGE, fractions containing the target protein were collected and pooled. Load the hydrophobic column HiTrap Phenyl HP as solution A for binding. Prepare low-salt eluent solution B (20mM Tris-HCl pH 8.0) for gradient elution. The eluted RBD was subjected to SDS gel electrophoresis to detect the target protein. The elution peaks containing the target protein obtained by crude purification by hydrophobic chromatography were combined, concentrated to 5 mL through a 10K cut-off concentration tube (Millipore), and then subjected to molecular sieve chromatography on a Superdex200 Hiload16 / 60 column (GE) for further purification of the t...
Embodiment 3
[0041] Embodiment 3: mouse immunization experiment
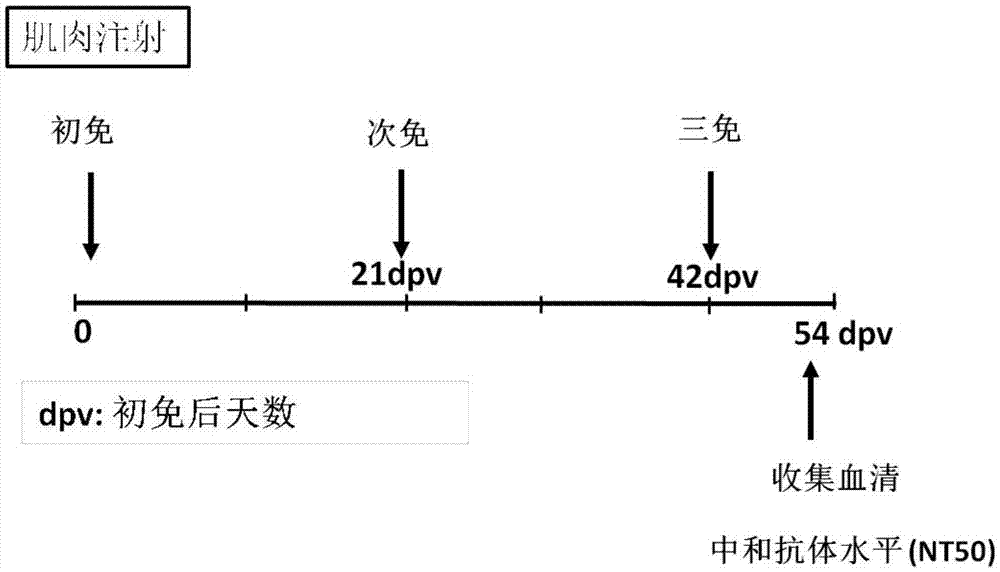
[0042] The MERS and SARS antigens obtained in Example 2 were diluted in physiological saline according to the method in Table 1, and emulsified in groups with adjuvants. Then 4-6 weeks old Balb / C mice were immunized in groups. immunization strategies such as image 3 , that is, through thigh muscle injection, each mouse received three vaccine immunizations on the 0th day, the 21st day, and the 42nd day respectively, with an inoculation volume of 100ul each time. On the 54th day (ie, the 12th day after the 3rd immunization), the tail blood was collected from the mice. The mouse serum was obtained by centrifuging at 3000 rpm for 10 minutes after standing for a period of time to allow the serum to separate out, and stored in a -20°C refrigerator for pseudovirus neutralization detection.
[0043] Table 1: Animal immunization groups
[0044] antigen or control Antigen content Adjuvant number of animals PB...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com