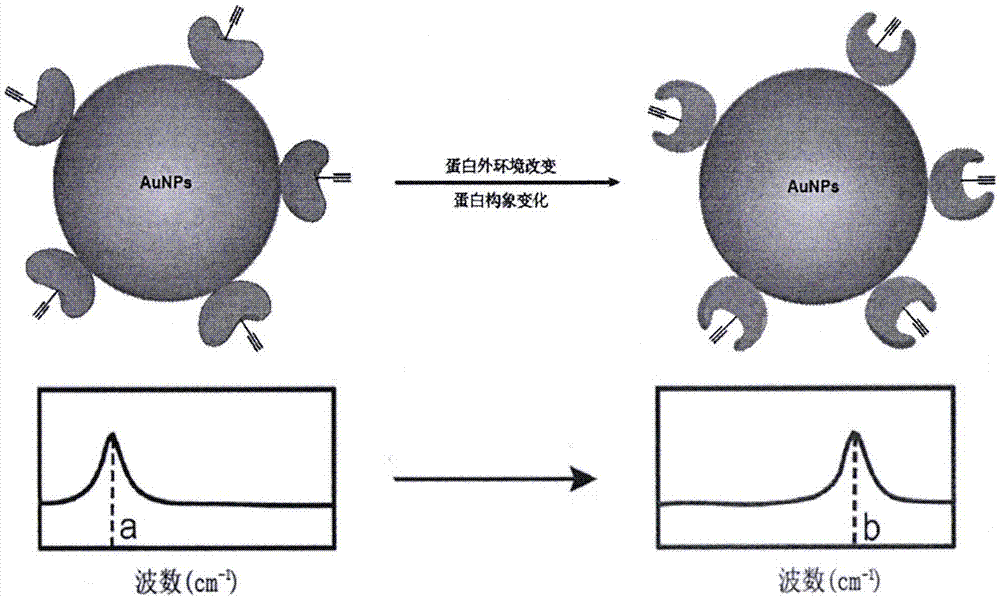
Biological orthogonal Raman in-situ detection method for conformation changes of protein
A bio-orthogonal and conformational change technology, which is applied in the detection of protein chemistry and conformational changes, can solve problems such as Raman spectral peak overlap, and achieve the effects of low interference of orthogonal Raman signals, simple data processing, and high sensitivity
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
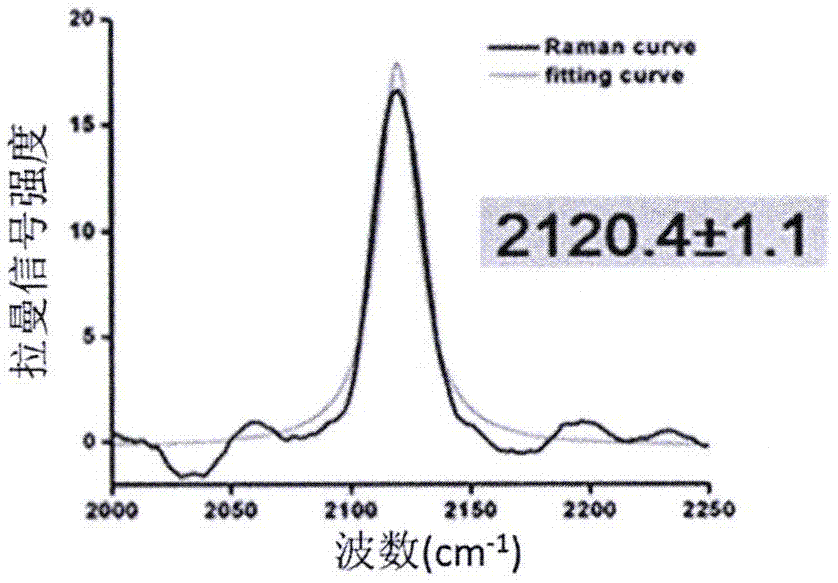
[0042] Example 1 Detection of conformational changes of alkyne-containing HdeA protein in solution at different pH
[0043] HdeA is a molecular chaperone protein in the bacterial membrane stroma. In the physiological environment of pH 7, HdeA exists in the form of homodimerization; while in the physiological environment of pH 2, HdeA depolymerizes to expose its active site and bind to its substrate protein. In the physiological environment of pH 2, HdeA is in a disordered state, and its structure and conformational changes cannot be studied by X-ray crystallography. Penk is a lysine derivative containing an alkyne group (as shown in the figure below), and the orthogonal Penk-MbPylRS / tRNA can be used to insert Penk into the TAG site of the protein.
[0044]
[0045] Synthesis of AuNPs: Gold nanoparticles (AuNPs) sol were prepared by sodium citrate reduction method, in which HAuCl 4 The final concentration of sodium citrate was 0.01% (w / w). In boiling ultrapure water, add ...
Embodiment 2
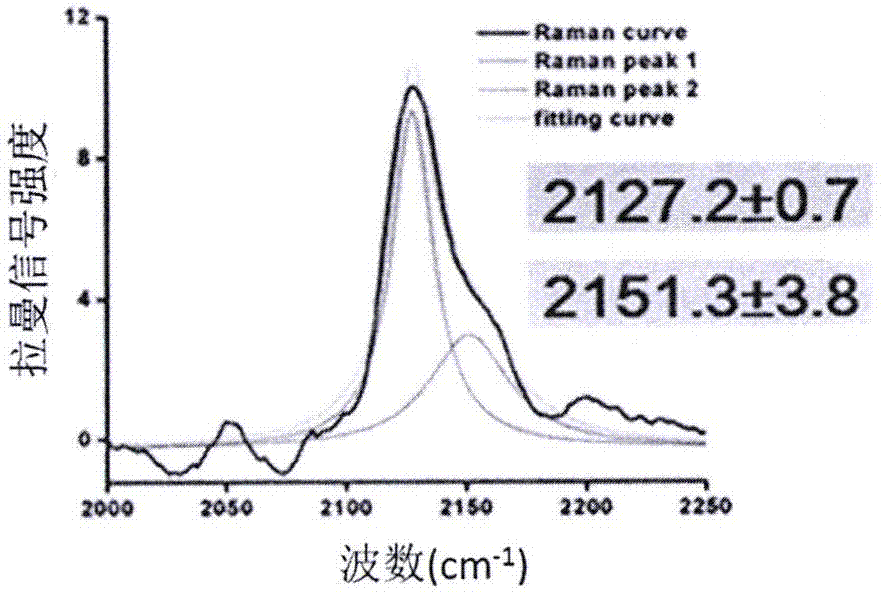
[0053] Example 2 Detection of conformational changes of cell surface alkyne-containing EGFR protein before and after EGF stimulation
[0054] EGFR is an epidermal growth factor receptor protein located on the surface of cell membranes. The EGFR signaling pathway plays an important role in physiological processes such as cell growth, proliferation and differentiation, and is closely related to a variety of cancers. EGFR will dimerize after combining with epidermal growth factor EGF, and EGF mainly exists in the form of monomer after being released.
[0055] Synthesis of AuNPs: Prepared by sodium citrate reduction method.
[0056] Expression of alkyne-containing EGFR in cells: the codons at positions 210, 250, 356 or 440 of the EGFR protein were mutated to TAG. In the presence of the unnatural amino acid Penk, will contain engineered EGFR and Penk-MbPylRS / tRNA Pyl CUA The plasmid of the gene sequence was transiently transformed into HEK293T cells. After 40 hours, the cells ...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com