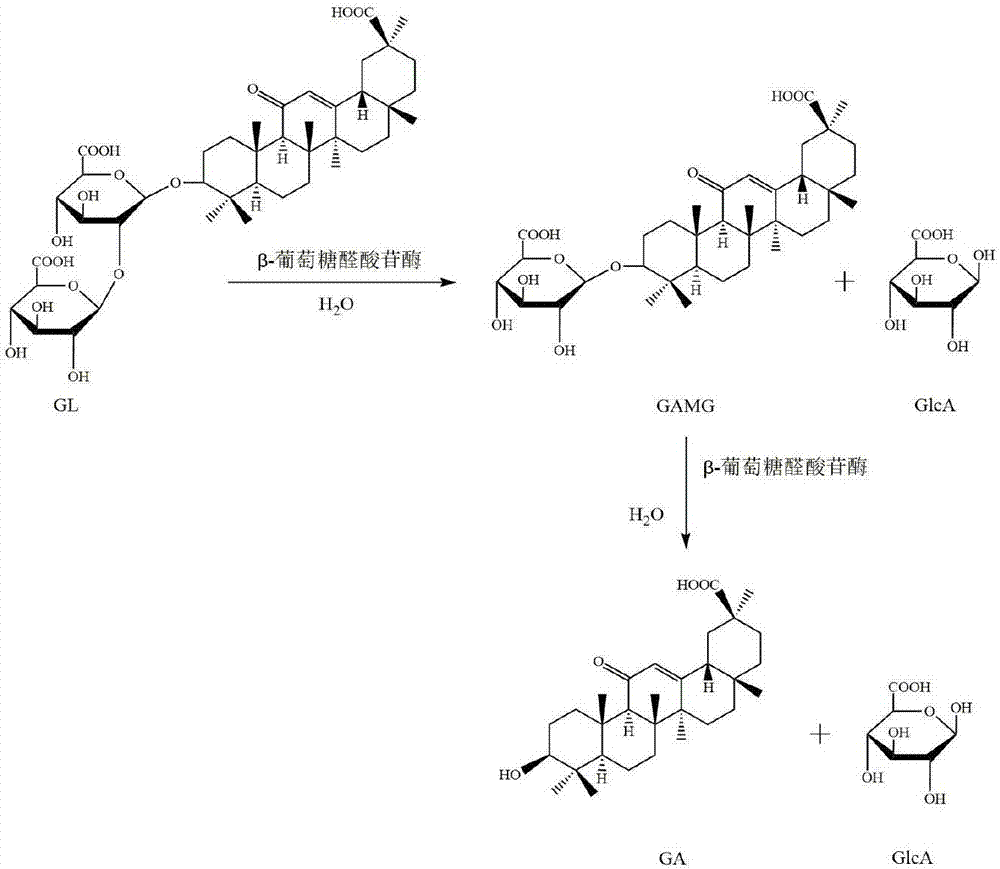
Beta-glucuronidase as well as gene and applications thereof
A technology of β-glucuronidase and glucose, applied in the field of genetic engineering, can solve the problems of low enzyme activity, poor substrate specificity, and low enzyme activity of β-glucuronidase
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 6
[0059] The monoammonium glycyrrhizinate in Example 6 has a purity of 75%, purchased from Xinjiang Tianshan Pharmaceutical Factory.
[0060] The enzyme activity definition of β-glucuronidase in the present invention: the amount of enzyme needed to generate 1 nmol GAMG per minute is an activity unit (U).
[0061] Among the present invention, the productive rate of GAMG is defined as:
[0062]
Embodiment 1
[0063] Example 1: Amplification of the T. pinophilum β-glucuronidase gene
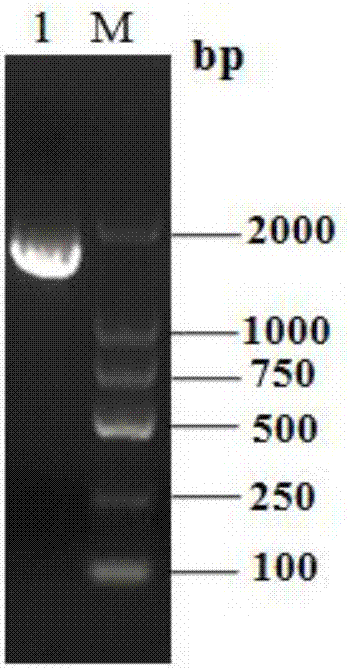
[0064] The total RNA of T. pinophilum Li-93 was extracted, reverse-transcribed into cDNA and used as a template, and the upstream primer Tpgus-F and downstream Tpgus-R were used to amplify the β-glucuronidase gene of T. pinophilum Tpgus, get the Tpgus gene fragment with a length of 1554bp, such as figure 2 shown. The gene fragment obtained above was connected with the cloning vector pMD19-T, transformed into Escherichia coli DH5α strain, the plasmid pMD19-T-Tpgus was extracted, and the Tpgus gene on the plasmid was sequenced and verified. The sequence was shown in SEQ ID No.1, and the results showed that it was correct .
Embodiment 2
[0065] Example 2: Construction of expression vector for T. pinophilum β-glucuronidase gene
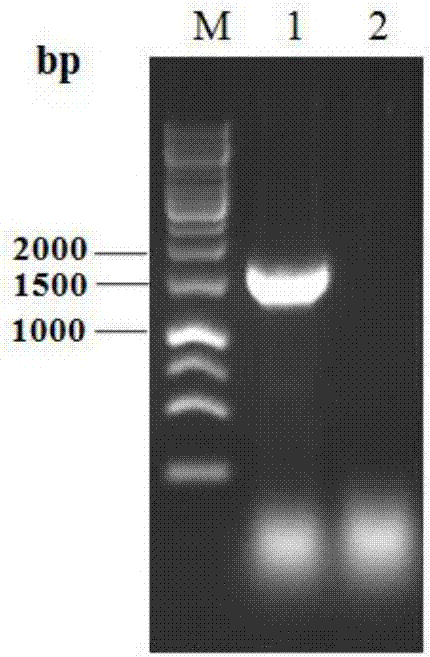
[0066] Taking pMD19-T-Tpgus in Example 1 as a template, using upstream primer Tpgus-F-KpnI and downstream primer Tpgus-R-NotI to amplify T. pinophilus beta-glucuronidase gene Tpgus, agarose electrophoresis Verify the amplification result, and cut the gel to recover the target fragment. After recovering the target fragment with a DNA gel recovery kit, perform double digestion with endonucleases KpnI and NotI, and then ligate with the pGAPZα plasmid that was also double-digested with endonucleases KpnI and NotI. The ligation product was transformed into Escherichia coli DH5α strain, and spread on the LB screening plate containing the antibiotic Zeocin. Colony PCR verification was performed using primers Tpgus-F-KpnI and 3'AOX. The positive clones were inoculated in LB liquid medium containing the antibiotic Zeocin, cultured overnight at 37°C, the plasmid was extracted, and the Tpgus ge...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com