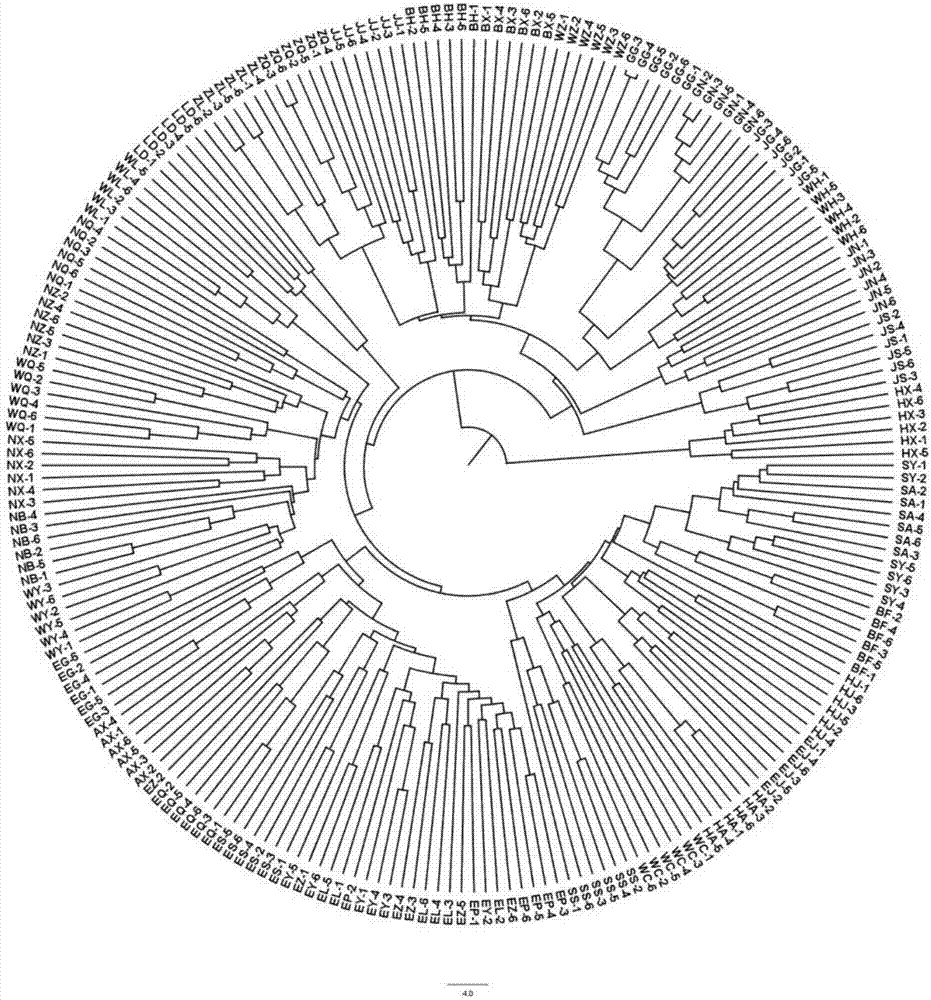
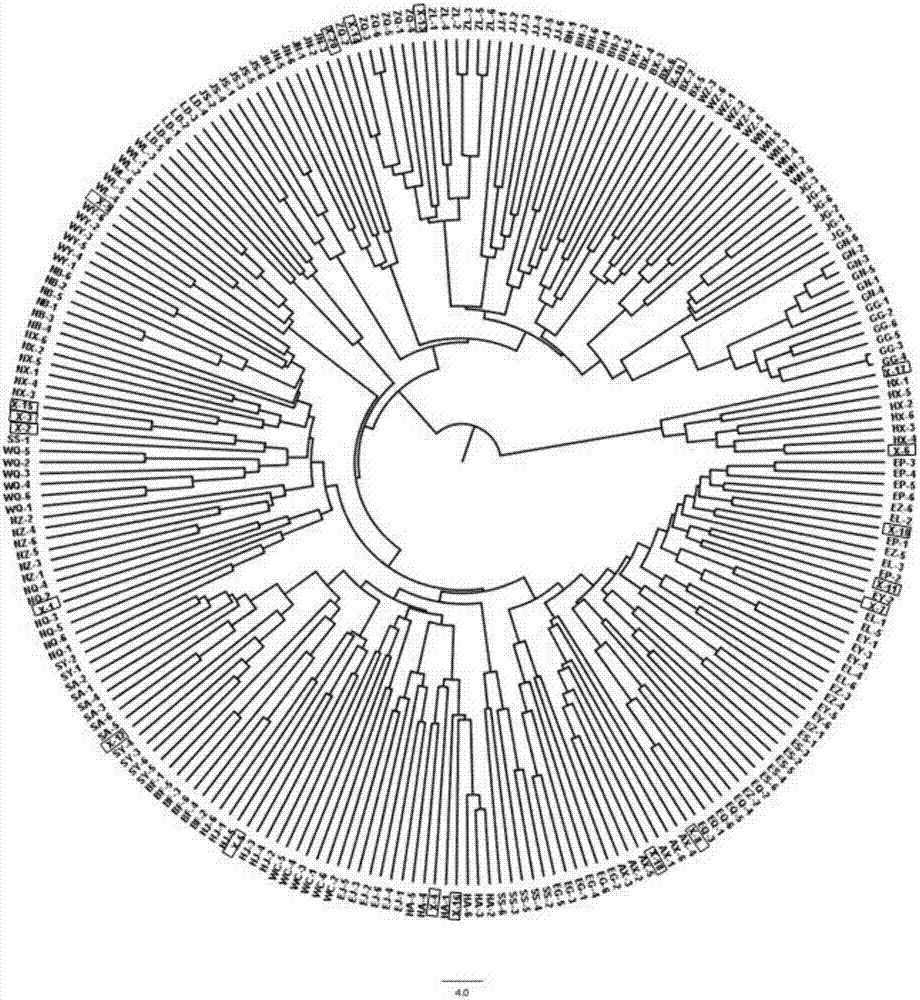
Method for identifying salvia miltiorrhiza germplasm resource
A technology of Salvia miltiorrhiza and germplasm is applied to the development and application of SSR primers in the identification of plant germplasm resources, which can solve the problems of inability to contain the genetic information of Salvia miltiorrhiza, the application defects of universal resource identification of experimental programs, etc., and achieve accurate results and identification. Easy-to-follow, intuitive results
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0041] Example 1. Construction of Salvia miltiorrhiza transcriptome database
[0042] (1) Use the RNAprep Pure Plant Kit (TIANGEN, BEIJING) kit to extract total RNA from fresh leaves of Salvia miltiorrhiza, and send it to a sequencing company for high-throughput transcriptome sequencing.
[0043] (2) Use the De novo assembly function of GENEIOUS 9.0.2 to splice short-read sequences into transcriptome frame data, take the longest transcript in each gene as Unigene, and establish a transcriptome database capable of searching for microsatellite sites .
Embodiment 2
[0044] Example 2. Development of microsatellite SSR primers
[0045] (1) Use the MISA (http: / / pgrc.ipk-gatersleben.de / misa / misa.html) program package to scan, identify, and locate the different types of microsatellite sites in the Unigene above; parameter settings: identify single bases 1, 2, 3, 4, 5, and 6 base repeating units in the SSR region, and the corresponding repetition times should be at least 10, 6, 5, 4, 3, and 3 , and the base distance between two adjacent SSRs is at least 100bp;
[0046] (2) Copy *.fasta, misa.pl and misa.ini to the same folder, run the command >mias.pl*.fasta in the Perl environment, and get *.fasta.misa and *.fasta after running Two files of .statistics, among which *.fasta.misa is used for subsequent primer design.
[0047] (3) Use the web version of BatchPrimer 3 to design SSR primers, set parameters: primer length 18-27bp, annealing temperature (Tm) 55-65°C, product length 50-500bp; display according to *.fasta.statistics and *.fasta.misa ...
Embodiment 3
[0049] Example 3. Genomic DNA Extraction
[0050] Using DNA PlantZol reagent, the modified CTAB method was used to extract the genomic DNA of the above 222 Danshen materials, quantified by NanoDrop 2000, diluted to 20ng / μl, and stored at 4°C or -20°C until use.
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com