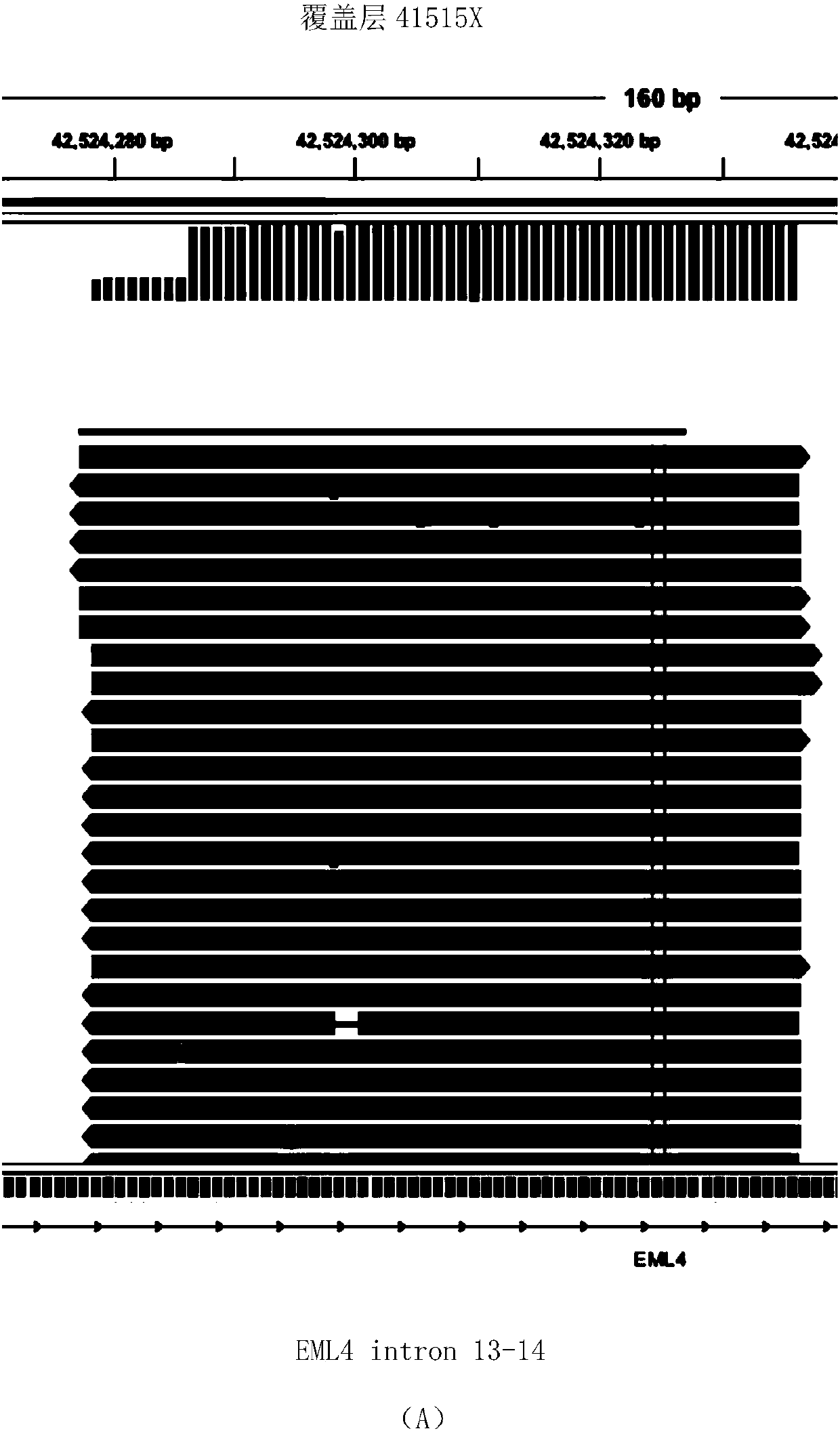
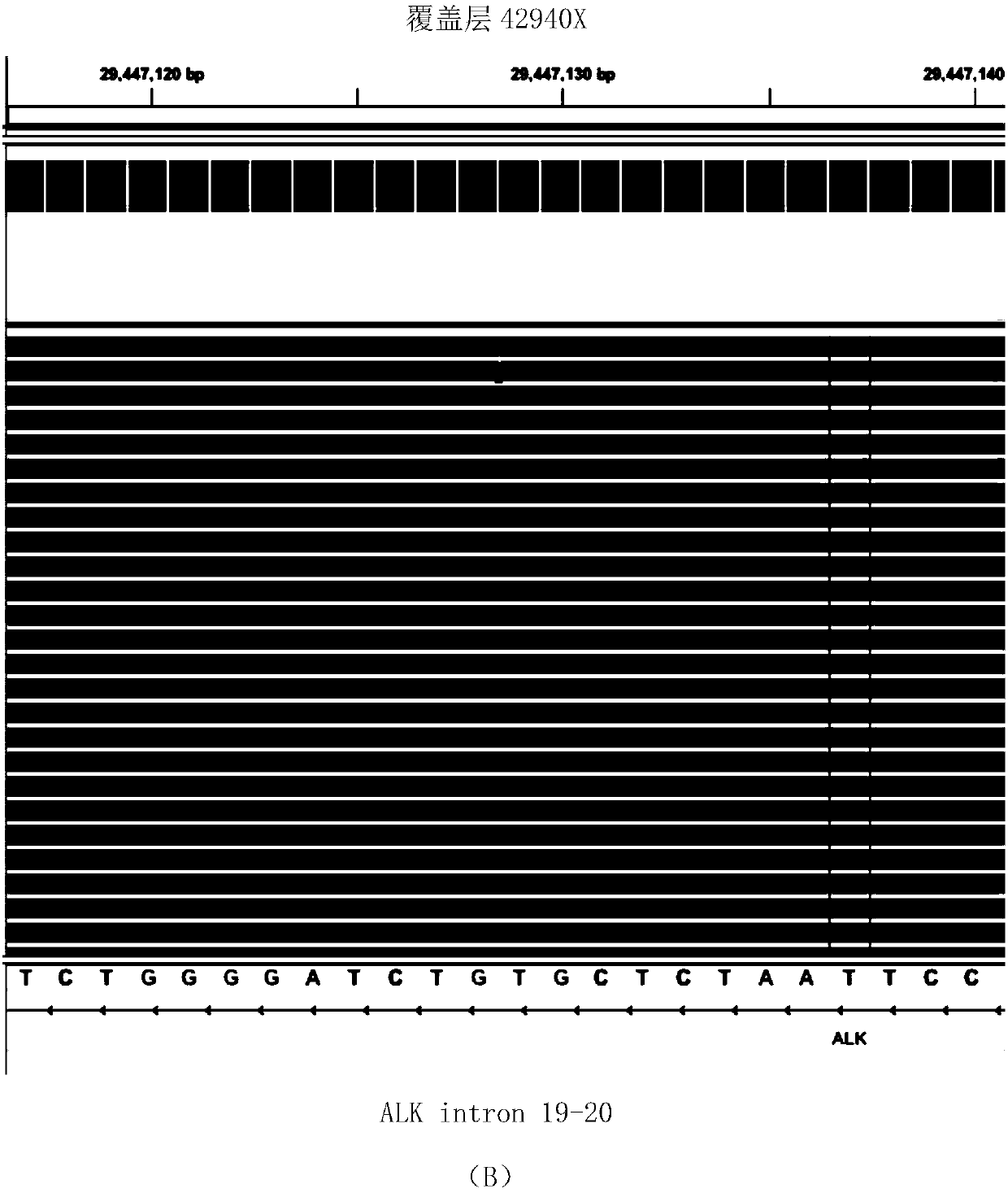
DNA-based fusion gene quantitative sequencing library construction, detection method and its application
A technology of fusion gene and DNA library, applied in recombinant DNA technology, microbial determination/inspection, DNA/RNA fragment, etc., can solve the problems of high operation difficulty, long operation time, high false positive rate, and short library construction time , high specificity, simple operation effect
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment Construction
[0040] In order to have a more specific understanding of the technical content, features, and effects of the present invention, the technical solution of the present invention will be described in further detail in conjunction with specific embodiments.
[0041] The equipment, experimental materials and reagents used in the examples are as follows:
[0042] 1. Instrument
[0043] AB ProFlex PCR instrument, IKA MS3Digital vortex oscillator, IVGN Qubit 3.0 fluorophotometer, IKA Mini G handheld centrifuge;
[0044] 2. Material
[0045] Rainin pipettes (specifications 10, 20, 200, 1000μl), AXYGEN universal filter tips (Universal FitFilter Tips, specifications 10, 20, 200, 1000μl), DYNAL DynaMag TM -2magnet magnetic stand, AmbionMagnetic Stand-96, AXYGEN 0.2ml transparent flat cover low adsorption PCR thin-walled tube, AXYGEN colorless low adsorption centrifuge tube (1.5ml, 2.0ml);
[0046] Three, reagent
[0047] Analytical pure absolute ethanol, IVGN dsDNA HS kit, NEB T4DNA ligase, NEB T4...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com