Bacteria engineered to treat diseases associated with hyperammonemia
A bacterial, genetically engineered technology, applied in the field of genetically engineered bacteria, capable of solving problems such as effective, reliable, and/or long-term unmet treatment
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0495] Example 1. ARG box mutations
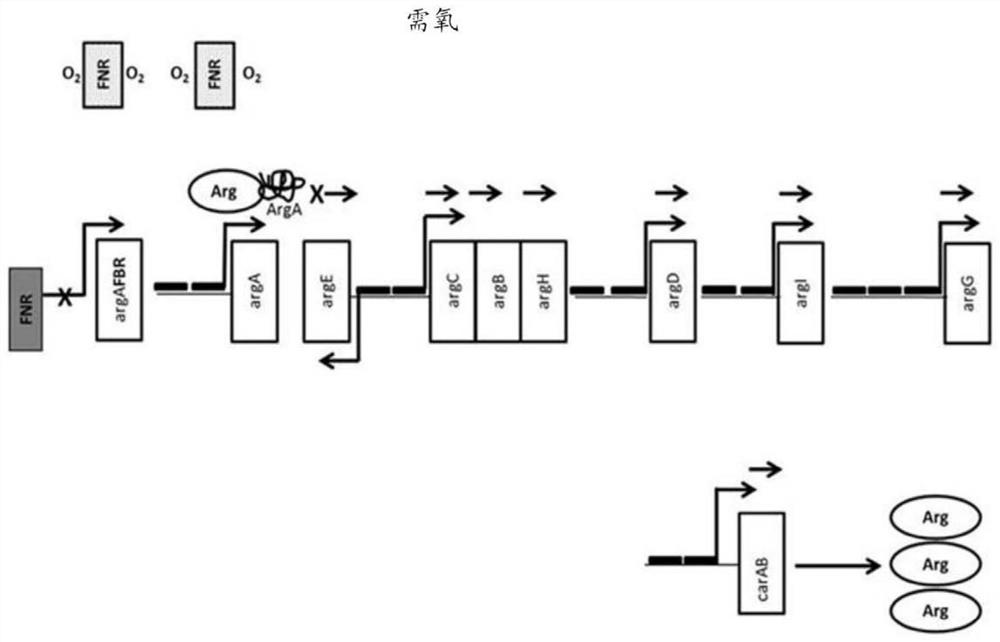
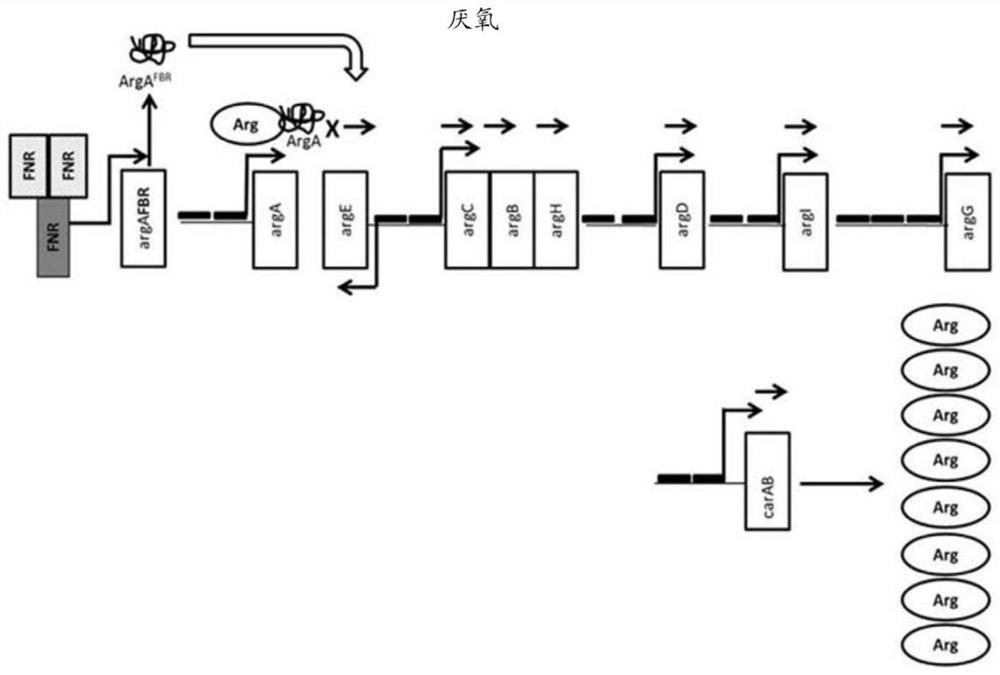
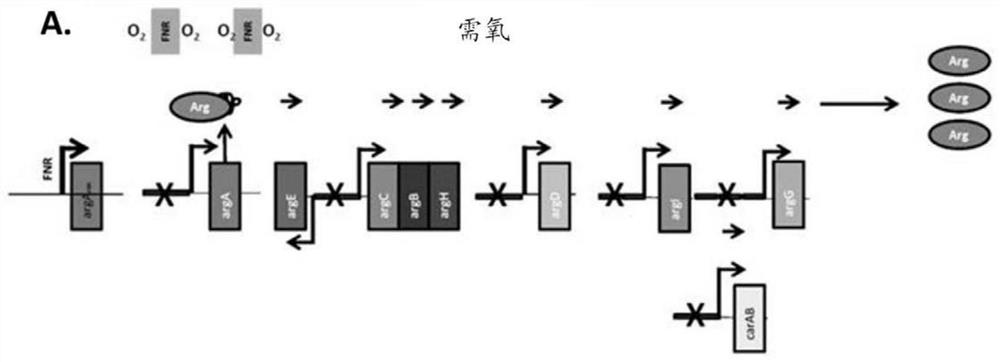
[0496] The wild-type genome sequence containing the ArgR binding site for each arginine biosynthesis operon in E. coli Nissle is shown in Figure 6 middle. Modifications to these sequences were designed according to the following parameters. For each wild-type sequence, the ARG box is shown in italics. The ARG box of the arginine regulon overlaps with the promoter region of each operon. quilt underlined Sequences represent RNA polymerase binding sites, and those sequences were not altered. Bases that are protected from DNA methylation during ArgR binding are highlighted, and bases that are protected from hydroxyl radical attack during ArgR binding are bolded. Highlighted and bolded bases are the primary targets of mutations for disrupting ArgR binding.
Embodiment 2
[0497] Embodiment 2.λred recombination
[0498] λred recombination is used to make chromosomal modifications such as ARG box mutations. λred is a program that uses the recombinase from bacteriophage λ to insert a piece of custom DNA into the E. coli chromosome. The pKD46 plasmid was transformed into E. coli Nissle host strain. E. coli Nissle cells were grown overnight in LB medium. Dilute the overnight culture 1:100 in 5 mL LB media and grow until it reaches OD 600 0.4-0.6. Pre-cool all tubes, solutions and cuvettes to 4°C. The E. coli cells were centrifuged at 2,000 rpm for 5 min at 4°C, the supernatant was removed, and the cells were resuspended in 1 mL of 4°C water. Escherichia coli was centrifuged at 2,000 rpm at 4°C for 5 min, the supernatant was removed, and the cells were resuspended in 0.5 mL of 4°C water. Escherichia coli was centrifuged at 2,000 rpm at 4°C for 5 min, the supernatant was removed, and the cells were resuspended in 0.1 mL of 4°C water. The electr...
Embodiment 3
[0504] Example 3. Transformation of Escherichia coli Nissle
[0505] The mutated ARG cassette construct was transformed into E. coli Nissle containing pKD46. Pre-cool all tubes, solutions and cuvettes to 4°C. Dilute the overnight culture 1:100 in 5 mL of LB medium containing ampicillin and grow until it reaches OD 600 is 0.1. Add 0.05 mL of 100X L-arabinose stock solution to induce pKD46 λred expression. Grow the culture until it reaches OD 600 0.4-0.6. The E. coli cells were centrifuged at 2,000 rpm for 5 min at 4°C, the supernatant was removed, and the cells were resuspended in 1 mL of 4°C water. Escherichia coli was centrifuged at 2,000 rpm at 4°C for 5 min, the supernatant was removed, and the cells were resuspended in 0.5 mL of 4°C water. Escherichia coli was centrifuged at 2,000 rpm at 4°C for 5 min, the supernatant was removed, and the cells were resuspended in 0.1 mL of 4°C water. Set the electroporator to 2.5 kV. 0.5 μg of the mutated ARG cassette construct wa...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com