Method for quickly detecting CYP2C19 genetic polymorphism and kit
A CYP2C19, gene polymorphism technology, applied in the field of molecular biology, can solve the problems of amplification (efficiency, specificity, selectivity, high detection cost, erroneous conclusions, etc., to save DNA extraction steps, high detection Specificity, fast detection effect
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0054] Embodiment 1: the processing of blood sample
[0055] In this example, 5 sets of blood were subjected to rapid processing after 40-fold dilution. The treatment solution is 20mM Tris-HCl pH8.0, 2mM EDTA, 2mg / ml proteinase K, mix 2ul of blood with 78ul of the treatment solution, mix thoroughly, and then keep warm at 56°C for 5min and 98°C for 2min, the crude sample is Processing complete.
Embodiment 2
[0056] Embodiment 2: the processing of saliva test sample
[0057] In this embodiment, the mucosal cells in the oral cavity were scraped with a special saliva test piece, part of the test piece was cut with scissors or the whole test piece was soaked, wherein the treatment solution was 20mM Tris-HCl pH8.0, 2mM EDTA, 2mg / ml proteinase K, immerse in the required volume, it is advisable to cover all the samples with the treatment solution, and shake vigorously for 1min, the crude sample is processed.
Embodiment 3
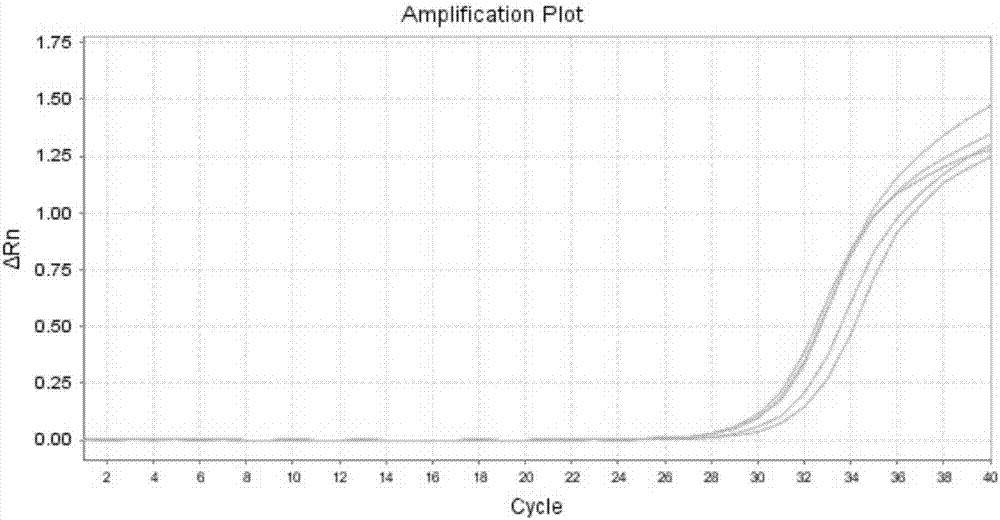
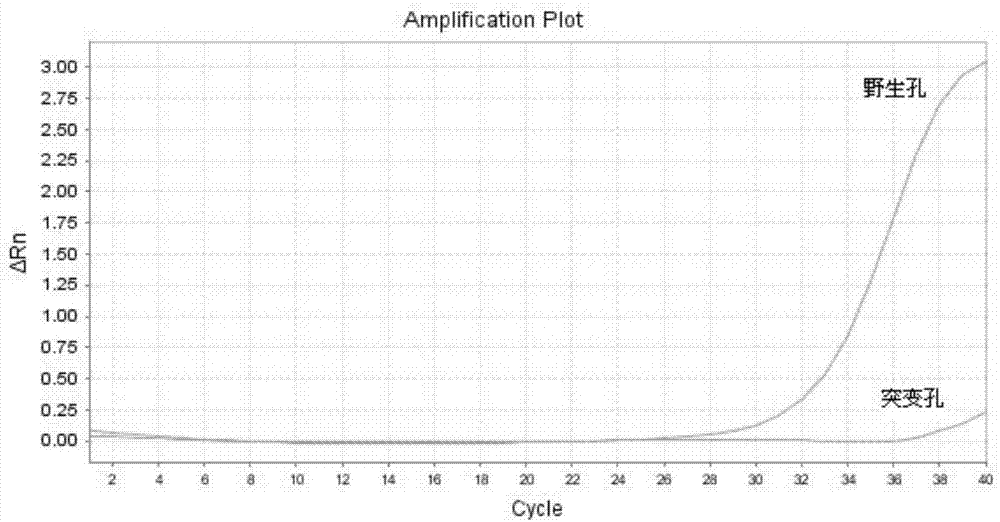
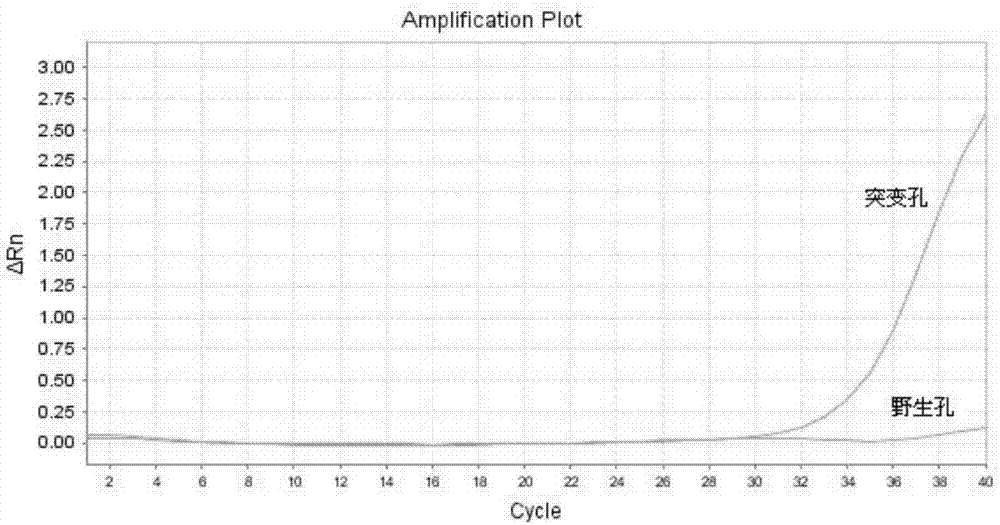
[0058] Embodiment 3: Rapid detection of blood samples
[0059] In this example, the blood is rapidly processed and the amplification of the internal reference gene is detected by using the method in Example 1 of the present invention.
[0060] The internal reference sequence is:
[0061] 5'-CGGACTGAAGGAGCTGCCCATGAGAAATTTACAGGGTGAGAGGCTGGGATGCCAAGGCTGGGGGTTCATAAATGCAGACAGCAGTTCCGATGGC-3'
[0062] Forward primer: 5'-CGGACTGAAGGAGCTGCC-3'
[0063] Reverse primer: 5'-GCCATCGGAACTGCTGTCTG-3'
[0064] Detection probe: 5'-Cy5-CCCCAGCCTTGGCATCCCA-BHQ2-3'
[0065] The PCR reaction system is 25 μl, each reaction system contains 2% glycerol, 200 μM each of dATP, dCTP, dGTP, and dTTP, 200 nM each of forward primer and reverse primer, 200 nM detection probe and 1 unit of hot-start Taq DNA polymerization enzyme. One unit refers to the amount of enzyme required to incorporate 10nmoldNTPs at 72°C for 30 minutes.
[0066] ABI 7500Fast fluorescent quantitative PCR instrument was used for ...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com