Optimized target object capturing system based on bacterial cell surface display system
A surface display system and bacterial cell technology, applied in the field of bioengineering, can solve the problems of cumbersome operation of the expression system, separation and acquisition of viral capsid proteins and receptors, etc., and achieve the effects of avoiding background interference, shortening the recovery cycle, and easy separation
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0074] Construction of prokaryotic expression plasmid vector for inducible surface display virus capsid protein:
[0075] The artificially synthesized inaQn-TB fusion gene fragment, whose sequence is shown in SEQ ID No: 5, was ligated into the prokaryotic cloning vector pUC57, and the ligated product was transformed into Escherichia coli DH5α by the heat shock method. Positive clones were screened on the LB plate, and the single colony picked out was cultured and the plasmid was extracted from it to obtain the recombinant plasmid pUC57-inaQn-TB.
[0076] Use Nco I / EcoR I and Bgl II / EcoR I to perform double enzyme digestion on pET28a-inaQn-P(GII.4), then use Nco I / Bgl II to double enzyme digest pUC57-inaQn-TB, and recover About 5.3kb, 960bp, 543bp DNA fragments. Put the above three fragments in T 4 Ligation is carried out under the action of DNA ligase, and the product after enzyme ligation is transformed into Escherichia coli DH5α. Positive clones were screened on LB plates...
Embodiment 2
[0078] Expression of prokaryotic expression plasmid of inducible surface display viral capsid protein and functional identification of surface display system:
[0079] The prokaryotic expression plasmid pET28-inaQn-TB-P (GII.4) prepared in Example 1 was transformed into Escherichia coli BL21, carrying the prokaryotic expression plasmid pET28-inaQn-TB-P (GII. 4) Escherichia coli BL21 was named IL-2.4P. Pick a single colony and inoculate it in LB medium containing 100 μg / mL kanamycin. After culturing at 37°C for 12h to 14h, inoculate it in a fresh medium with 1.0% inoculation amount, and continue shaking culture until OD 600nmWhen it is 0.6, add IPTG with a final concentration of 0.4mmol / L, and continue shaking culture at 26°C for 16h. Centrifuge at 5000rpm for 10min at 4°C to collect the bacteria, then resuspend the bacteria in sterile PBS buffer with pH 7.2, OD 600nm Adjust to 1.0 and store at 4°C for later use.
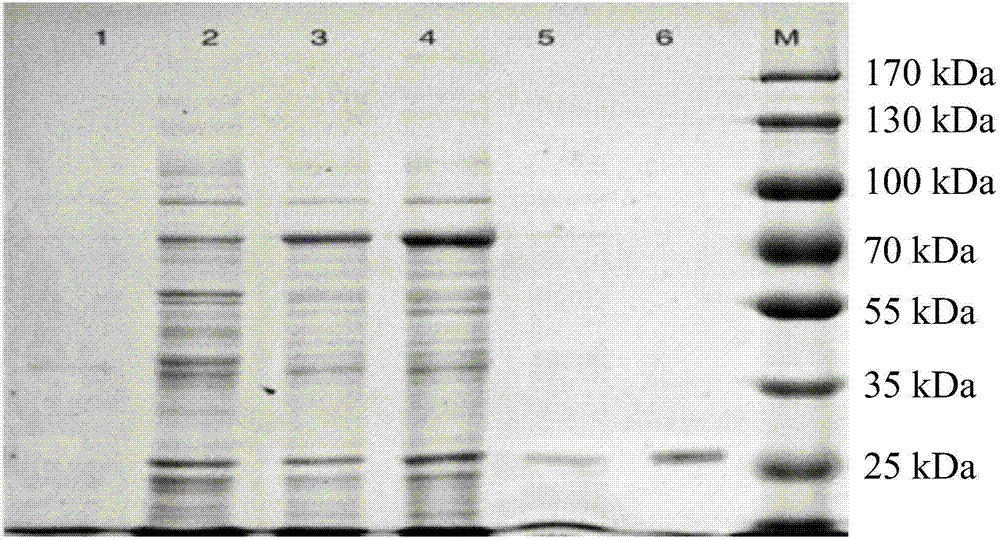
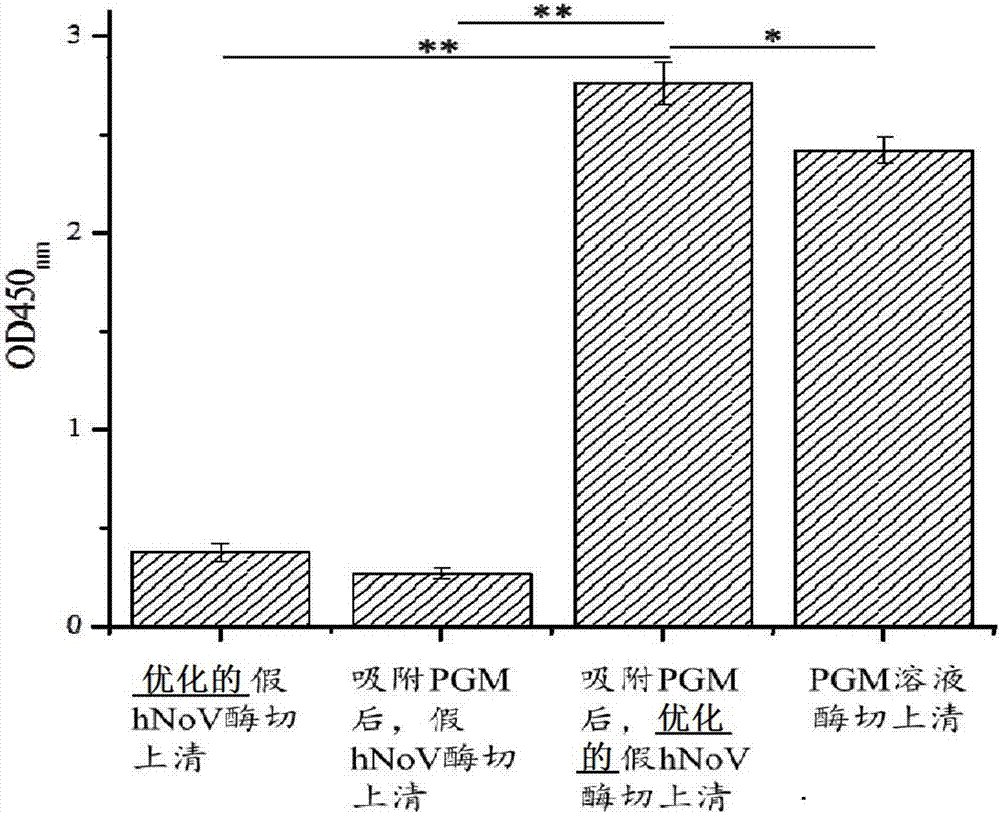
[0080] Such as figure 1 As shown, by SDS-PAGE detection of ...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com