Accurate identification method of nucleic acid
A recognition method and nucleic acid technology, applied in the field of biometrics, can solve problems such as difficult to achieve high-resolution imaging, decrease in specific hybridization intensity, and hinder resolution, achieve stable and controllable quality of nucleic acid coatings and probes, and improve specific hybridization Intensity, reducing the effect of non-specific binding
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0114] The preparation of embodiment 1 cell sample (I)
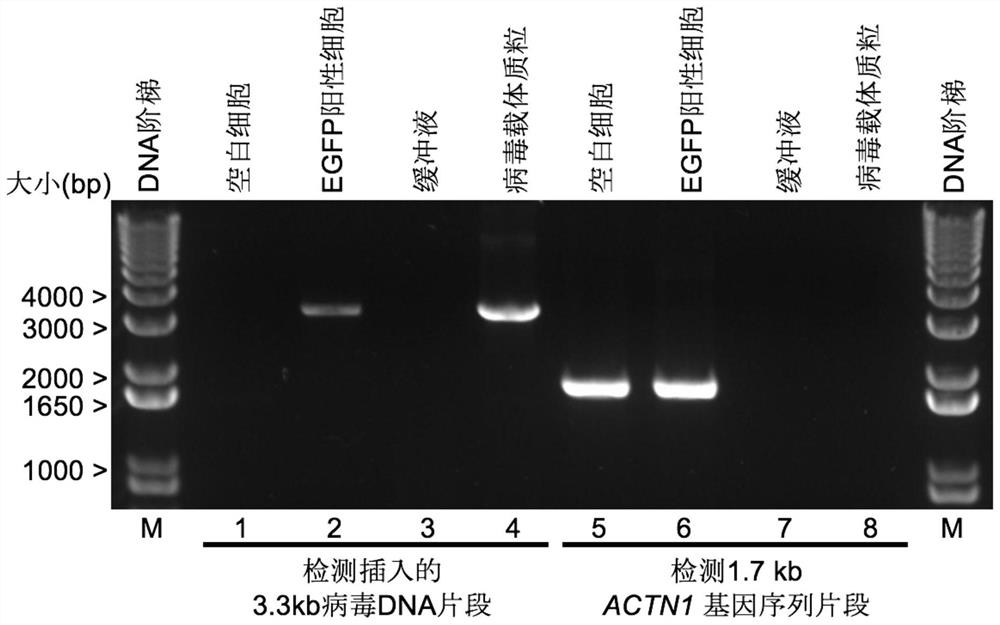
[0115] Human SK-N-SH cells were infected with the engineered pLL3.7-based lentiviral vector. After infection, cells positive for EGFP expression were screened by flow cytometry (BD FACSAria SORP Cell Sorter). In MEM medium (Gibco) containing 10% fetal bovine serum, on coverslips (Fisherbrand TM Coverglass for Growth TM Cover Glasses, Cat. No. 12-545-82) were cultured on the above-mentioned EGFP-positive cells (positive cells) or human SK-N-SH cells not infected by lentivirus (negative control).
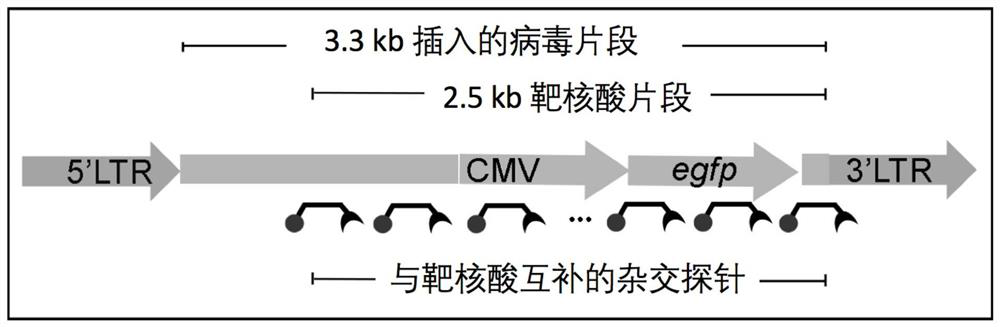
[0116] In the positive cells, a viral RNA fragment with a length of about 3.3 kb of the above-mentioned lentiviral vector was integrated into the genome of the SK-N-SH cell through reverse transcription. The integrated 3.3 kb fragment (SEQ ID NO:30) contained the sequence encoding EGFP expressed from the CMV promoter. A fragment of about 2.5 kb in length among the fragments of 3.3 kb is identified as an exemplary target nu...
Embodiment 2
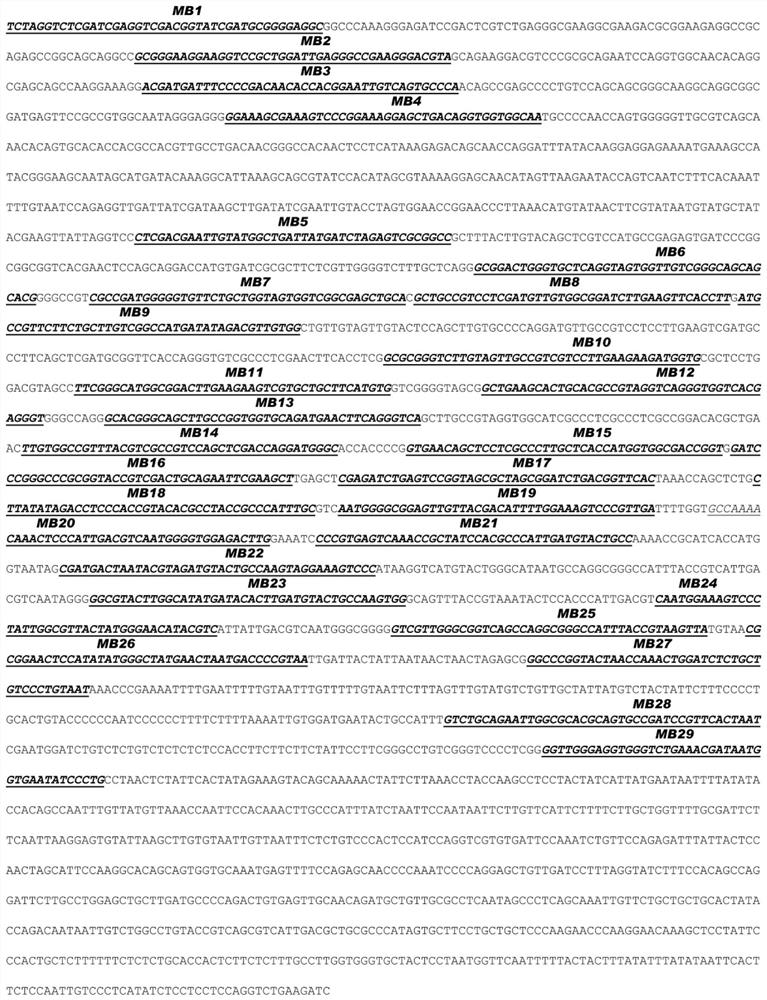
[0117] Design and synthesis of embodiment 2 hybridization probes MB1 to MB29
[0118] The exemplary hybridization probe provided in this example is a 56nt single-stranded oligonucleotide comprising a 42nt loop structure and 7nt arm structures on both sides of the loop structure. One end of the hybridization probe is conjugated with Alexa-647 as a fluorophore, and the other end is conjugated with BHQ3 as a quencher.
[0119] 29 hybridization probes were designed, denoted as MB1 to MB29, and synthesized by Life Technology. The sequences of these 29 hybridization probes are recorded in the sequence listing (SEQ ID NO: 1-29). The sequence of the loop structure is reverse complementary to a part of the antisense strand of the target nucleic acid fragment in Example 1 (that is, identical to a part of the sense strand of the target nucleic acid fragment), and 29 hybridization probes can pass through their respective loop structures The target nucleic acid fragments are labeled alon...
Embodiment 3
[0124] Embodiment 3 Hybridization probes MB1 to MB29 are specifically combined with target nucleic acid
[0125] In a buffer containing 50 mM NaCl, 1 mM EDTA, 10 mM Tris (pH 7.4), dissolve the hybridization probe, the oligonucleotide complementary to the hybridization probe (CS), and the oligonucleotide non-complementary to the hybridization probe sequence acid (NCS).
[0126] 80 nM hybridization probes (MB1 to MB29) and corresponding CS (1600 nM) or NCS (1600 nM) were reacted in 2×SSC, 50% formamide hybridization solution for 30 min at room temperature. Thereafter, fluorescence emission readings were taken at 665 nm using a spectrofluorometer excited with a 647 nm laser. Readouts for each hybridization probe are shown in Figure 3a . For each hybridization probe, the fluorescence emission readout after co-reaction with CS was significantly higher than that after co-reaction with NCS, indicating the specific binding of each hybridization probe to the target sequence.
[01...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com